† These authors contributed equally.
Academic Editor: Graham Pawelec
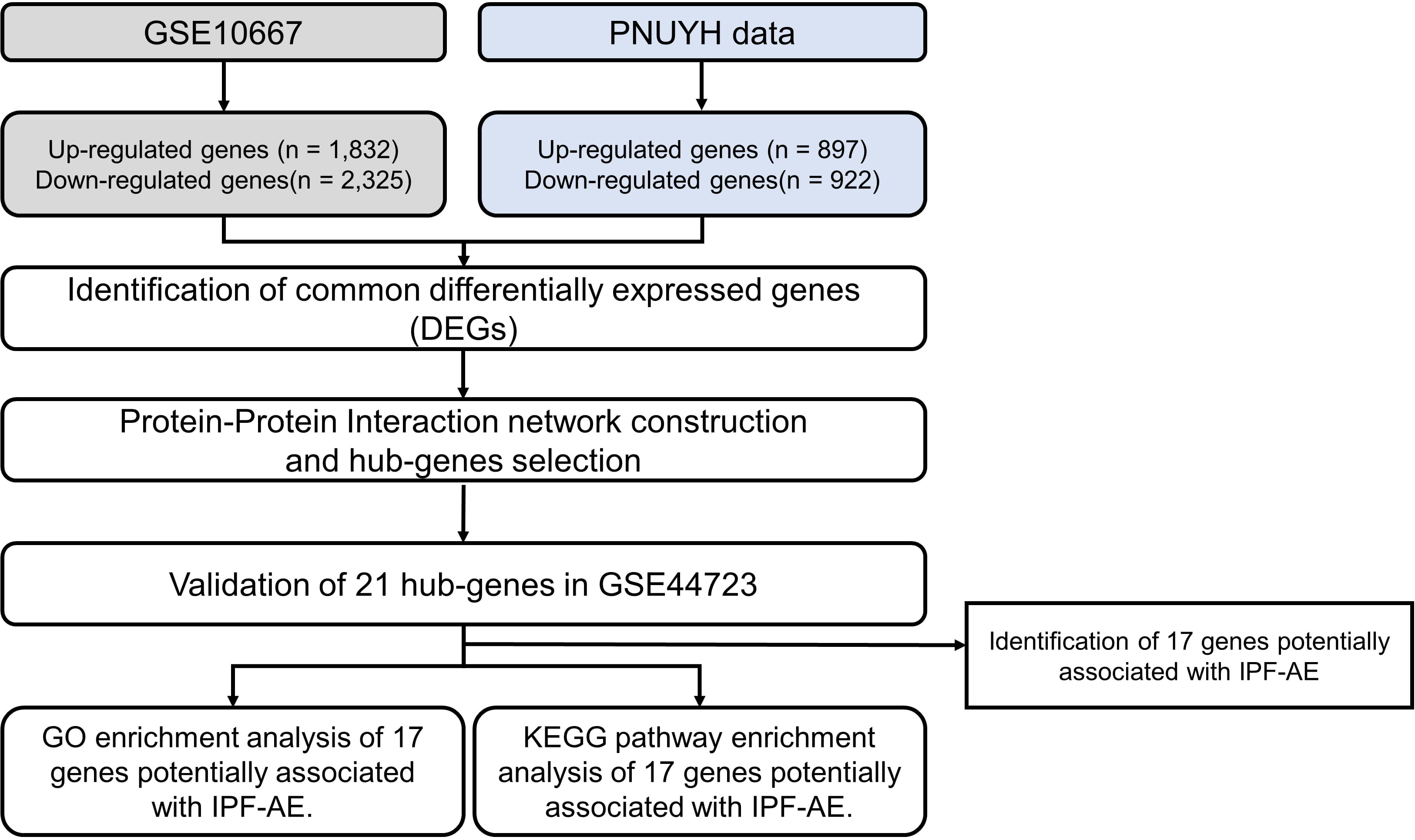
Introduction: The molecular mechanisms underlying acute exacerbations (AEs) of idiopathic pulmonary fibrosis (IPF) are poorly understood. To understand the gene expression patterns of the AEs of IPF, we studied gene expression profiling of AEs of IPF. Methods: The GEO datasets included in this study are GSE44723 and GSE10667, and in-house RNA-seq data were used. DEG analysis used the limma package, and the STRING database was used to construct the protein-protein interaction (PPI) network, and its functional role was investigated through gene ontology analysis. Results: The results of DEG analysis indicated 76 upregulated and 135 downregulated genes associated with an AE of IPF compared to stable IPF. The PPI network included three core modules containing 24 of the 211 DEGs. Eleven upregulated and six downregulated genes were evident in AEs of IPF compared with stable IPF after validation. The upregulated genes were associated with cell division. The downregulated genes were related to skeletal muscle differentiation and development. Conclusion: In previous studies, 17 genes were strongly associated with cell proliferation in various cell types. In particular, cyclin A2 (CCNA2) was overexpressed in the alveolar epithelium of the lungs presenting AEs of IPF. Aside from the previously described CCNA2, this study reveals 16 genes associated with AEs of IPF. This data could indicate new therapeutic targets and potential biomarkers for the AEs of IPF.