† These authors contributed equally.
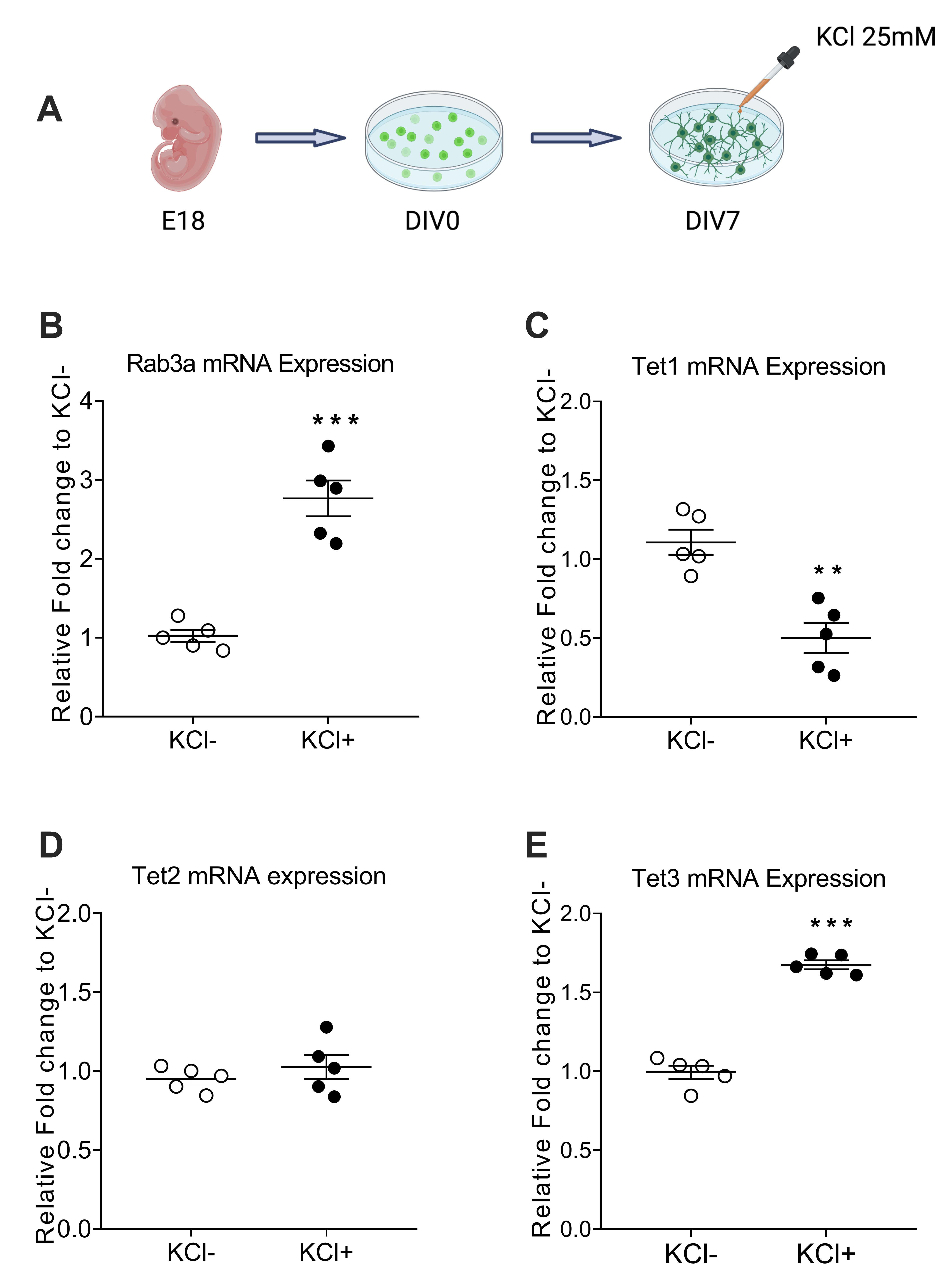
Rab3a, a subtype protein in the Rab3 family amongst the small G proteins, is closely associated with the learning and memory formation process. Various neuronal stimuli can induce the expression of Rab3a; however, how DNA modification is involved in regulating its expression is not fully understood. Ten-eleven translocation (TET) proteins can oxidate methylcytosine to hydroxymethylcytosine, which can further activate gene expression. Previous studies reported that TET-mediated regulation of 5hmC induced by learning is involved in neuronal activation. However, whether Tet protein regulates Rab3a is unknown. To understand the role of TET-mediated 5hmC on Rab3a in neuronal activation, we adopted a KCl-induced depolarization protocol in cultured primary cortical neurons to mimic neuronal activity in vitro. After KCl treatment, Rab3a and Tet3 mRNA expression were induced. Moreover, we observed a decrease in the methylation level and an increase of hydroxymethylation level surrounding the CpG island near the transcription start site of Rab3a. Furthermore, recently, Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) has proven powerful in identifying open chromatin in the genome of various eukaryotes. Using FAIRE-qPCR, we observed a euchromatin state and the increased occupancy of Tet3, H3K4me3, and H3K27ac at the promoter region of Rab3a after KCl treatment. Finally, by using shRNA to knockdown Tet3 prior KCl treatment, all changes mentioned above vanished. Thus, our findings elucidated that the neuronal activity-induced accumulation of hydroxymethylation, which Tet3 mediates, can introduce an active and permissive chromatin structure at Rab3a promoter and lead to the induction of Rab3a mRNA expression.