MicroRNAs (miRNAs) are endogenous, small non-coding RNA molecules that play important regulatory roles in numerous biological processes, cellular pathways and networks. They do so by targeting specific mRNAs, directly degrading them and/or preventing their translation into proteins. The impaired function of miRNAs results in aberrant gene expression that promotes abnormal cell growth and differentiation. miRNAs, located at fragile sites or cancer associated regions of the genome, act as either tumor suppressor or tumor promoters. miRNAs also play significant role in aging and in various diseases. Despite the fact that profiling of miRNA expression is considered an important tool for diagnostic and therapeutic purposes, such analysis has not yet been adopted in routine clinical care practices. Here, we highlight advances and challenges in research on cancer and aging in relation to miRNAs.
A microRNA (miRNA) is of about 22 nucleotide non-coding RNA that regulates gene expression by sequence specific base pairing with the target mRNA in the 3’ UTR. Thereby, they induce direct mRNA degradation or translational inhibition (1). miRNAs have been shown to play crucial role(s) in numerous biological processes, cellular pathways and networks. The impaired function of miRNA results in aberrant gene expression that promotes abnormal cell growth and differentiation. miRNAs are located at fragile sites or cancer associated regions of the genome. They act as either tumor suppressor or tumor promoting miRNAs (oncomiRs), thereby, involving themselves in tumorigenesis and tumor progression. miRNAs also found to have immense role in aging and its associated events in cells. The quantitative and qualitative assessment of miRNAs expression has also indicated remarkable changes in various diseases other than cancer and aged states. Given the involvement of miRNAs in cancer and aging, they have become an area of intense research focus in recent years.
The first miRNA was discovered in 1993 in the Caenorhabditis elegans and, it was lin-4. It is a non-coding miRNA containing complementary sequence in the 3’ untranslated region (UTR) (2). Ever since this discovery, extensive research has been carried out on several miRNA molecules to get an insight on their role in various cellular processes. More than a decade ago, Bartel (2004) estimated that about 30% of human genes are regulated by miRNA and, later in 2007 (3), Perera and Ray (2007) identified that mRNA are the targets of 20-30% of miRNA (4). As miRNAs share seed sequences and with the presence of overlapping targets, a single miRNA targets many different mRNAs and also different miRNAs can target the same mRNA (5). It indicates that miRNAs act as key regulators in all biological pathways in multicellular organisms including mammals. Under normal physiological conditions, miRNAs regulate biological processes including cell differentiation, proliferation and apoptosis in cancer cells. They play a vital role in regulating gene expression at post transcriptional level and also in establishing cell’s identity. Therefore, impaired miRNA function results in aberrant gene expression that promotes abnormal cell growth and differentiation (6). After the characterization of miR-15 and miR-16 in chronic lymphocytic leukemia in the year 2002, miRNA was considered as hallmark for cancer. Emerging evidences suggest that miRNA are involved in cancer development and progression by variety of stimuli including transcriptional activation, epigenetic modifications, genomic amplification or deletion, cellular stress and inflammatory stimuli. Such activities of miRNAs eventually influence the most of biological process including cell proliferation, DNA repair, DNA methylation, apoptosis and pro-inflammatory or anti-inflammatory signals (7). MicroRNAs either contribute to modulate or directly modulate the cancer by inhibiting the expression of tumor suppressors or oncogenes. When they contribute for the expression of oncogenes, they are called as oncogenic miRNAs (onco-miRNA). Generally, onco-miRNAs are over expressed in cancers while tumor suppressive miRNAs are down regulated. When the functions of these onco-miRs or tumor suppressors are reversed, proliferation, metastasis and survival of cancer cells can be significantly reversed (8). On the other hand, the role of miRNAs are also been elegantly studies and evidenced for progression of aging and its associated complications. The present review is a collection of literature to draw intricate relation among cancer, aging and their control by miRNA. Although miRNAs have intricate and provoking role with many metabolic disorder and diseases, they have not been considered as a routine clinical marker. Thus, profiling of miRNA expression can be an important tool for diagnostics and treatment of disease. Since, miRNAs analysis until have not been adopted neither as diagnostic markers nor as clinical targets in routine clinical practices, the objective of this article was to highlight advances and challenges in research on cancer and aging in relation to miRNA in order to make correlation among miRNA, aging and cancer.
miRNAs are evolutionarily conserved and found to be located in the introns or exons or between genes and is synthesized primarily from primiRNA. PrimiRNA is a long and primary transcripts of several kilobase pairs and are transcribed by RNA polymerase II (9). Drosha, an RNase III enzyme, cleaves this long primiRNA into 60 – 70 nucleotide precursor miRNA, known as pre-miRNA (10, 11), which forms a characteristic hairpin double-strand structure. It is exported from the nucleus to the cytoplasm by Exportin-5 (12). Then it binds to RNase Dicer, a second RNase III enzyme and RISC (RNA – Induced Silencing Complex). RISC is composed of Transactivation Responsive RNA Binding Protein (TRBP) and Argonaute 2 (Ago 2). Initially, 12 nucleotides are removed from the pre-miRNA at 3’ end by Ago 2. Further, dicer cleaves the intermediate to produce a mature 22 nucleotide miRNA (13). The mature strand is retained by RISC while the inactive or “passenger” strand is removed and degraded (3, 13, 14, 15). The mature miRNA recognizes its target mRNA by complementary base pairing in the 3’ UTR (16, 17). The detailed pathway is described elsewhere but one of the very common pathways of biogenesis of miRNA is demonstrated in Figure 1.
 Figure 1
Figure 1Biogenesis miRNA.
MicroRNAs harbor attractive features ranging from translation to clinical practices, such as simple extraction, resistance to molecular degradation, and accurate, reproducible quantification using quantitative real-time reverse transcription PCR (qRT-PCR) and other analytical methods. Combined with their potential to improve cancer diagnosis, classification, prognosis and therapies, miRNAs have garnered significant research attention in recent years. For instance, with the identification of up-regulated miR-141 in prostate cancer patients, circulating serum miRNAs were speculated as a possible non-invasive diagnostic tool. miRNA profiling may also facilitate better classification of tumors into treatment subgroups, especially for poorly differentiated tumors that are difficult to be defined by histology. A number of prognostic miRNA signatures have been generated and validated in various cancers (18). This part of review summarizes the role of miRNA in the diagnosis and prognosis of various human cancers. A consolidated data on the role of miRNAs and their target in different organs is presented in Table 1.

|
Breast cancer is the second most cancer diagnosed worldwide, affecting 1.3 million women per year and accounts for 23% of all cancer cases. It is commonly categorized into one of four main subtypes such as luminal A, luminal B, human epidermal growth factor receptor 2 (HER2) positive and basal. More than 2000 miRNAs have been identified and some miRNAs have been shown to play crucial role in numerous biological processes, cellular pathways and networks. miRNA regulates major cellular functions such as development, differentiation, growth, metabolism, survival, motility and proliferation. MicroRNAs may act as either tumor suppressor or as onco-miRNA in breast cancer too (19, 20). Yu et al. demonstrated that miR-17/20 cluster modulates cell migration and invasion of nearby cells in breast cancer by inhibiting cyclin D1. They have also identified that miR-17/20 regulates metastasis by heterotypic secreted signal. Later in 2014, they found that miR-17/20 is able to trigger apoptosis by activating p53 that subsequently promotes degradation of cell survival oncoprotein, Akt (21, 22) Figure 2). Similarly, in comparison with healthy controls, miR-205 is down regulated in breast cancer samples whose ectopic expression suppresses proliferation and finally, it increases tumor cell apoptosis (23).
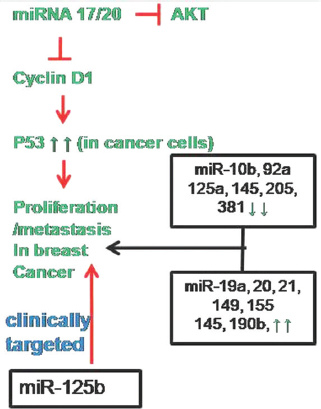 Figure 2
Figure 2Involvement of miRNA in breast cancer pathway. Out of many miRNA regulates breast cancer, miRNA-125b is proposed to be clinically targeted as marker.
Several miRNAs are screened for their role in breast cancer. Iorio et al. demonstrated the expression pattern of 15 miRNAs in 76 breast cancer samples against 10 normal breast samples in which five miRNAs (miR10b, miR-125b, miR-145, miR21 and miR-155) have been found to express differentially. Among those five miRNAs, miR-10b, miR-125b, miR-145 levels were seemed to be down regulated while miR-21 and miR-155 were noticed to be up regulated. Thus, they might possibly act as either tumor suppressor or oncogene, respectively (24). Schwarzenbach et al. have screened serum samples collected from102 pre-operative and post-operative patients with early breast cancer along with 53 healthy women for differential miRNA expression pattern (25). The miRNA signature showed that miR-20 and miR-21 are expressed notably higher in patient samples than in healthy individuals while coherent observation revealed malignant and benign tumor can be better distinguished with varied miR-214 expression (25). Another study on miRNA profile demonstrated the relative expression of 10 miRNAs (miR-106b, miR-125b, miR-17, miR-185, miR-21, miR-558, miR-625, miR-665, miR-92a, and miR-93) in 48 tissues and 100 serum samples from breast cancer patients combined with a set of 20 samples from healthy women. It was identified that there was a decrease in miR-92a levels but on the other hand, an increase in miR-21 level was noted, whose expression had remarkable association with tumor size and positive lymph node status (26). Similarly, signature pattern of miR-155 showed 2 folds increase whereas other miRNAs such as let-7b, miR-381, miR-10b, miR-125a-5p, miR-335, miR-205 and miR-145 were down regulated in breast cancer tissue samples. Detailed analysis on miR-155 confirmed its relationship with respect to clinical stage, molecular type, ki-67 and p53 status (23). Subsequently, miRNAs are focused for their potency to act as biomarkers in diagnosis and prognosis to improve cancer management.
Metastasis, a major obstacle for treatment of cancer, is an intricate, multistep process involving escape from the primary tumor site, local invasion, intravasation, and survival in the systemic circulation, extravasation into distant organs and finally successful colonization and outgrowth at the secondary sites. Recently, many researchers have shown interest in identifying miRNAs to use them as biomarkers for metastatic breast cancer. Chan and coworkers identified a novel metastasis suppressor miRNA i.e. miR149. It regulates GIT1 in vitro and in vivo and therefore, is proposed to be used as promising biomarkers as well as for anti-metastasic molecule in the treatment of breast cancer (27). Similarly, miR-19a is seen at high level in the serum of breast cancer patients which might represent as a biomarker for the patients with metastasis HER2+ breast cancer (28). MicroRNA-621 enhances chemo-sensitivity of breast cancer cells to PTX/CBP chemotherapy by suppressing FBXO11-dependent inhibition of p53 while, miR-190b is shown to be highly up regulated in hormone-dependent breast cancer. Thus, miR-190b acts as a new biomarker in hormone dependent breast cancers. Considering the widespread use of aromatase inhibitors (AIs) in endocrine therapy as a first-line treatment for postmenopausal estrogen receptor α–positive breast cancer patients, miR-125b is found to be a new candidate therapeutic target in AI-resistant breast cancers (29-31). As discussed, due to obvious connection with carcinogenesis, miRNA have been proved to be promising candidate for diagnostic and therapeutic potential. Even though, several miRNAs represent a novel class of potential biomarkers, still it has to be firmly validated so that they could be employed to be used in clinical practices.
Colorectal cancer (CRC) is a heterogeneous cancer including both colon cancer and rectal cancer. More than 1 million new cancers related cases and 600,000 deaths are expected to occur every year. CRC is curable by chemotherapy or surgery due to its confined growth within the wall of colon but cancers with strong metastasis activity are often lethal. Given the invasive nature and expensive cost of current screening methods of CRC diagnosis, it is difficult to efficiently detect CRC under early stages. Therefore, discovery of an effective, noninvasive blood-based molecular CRC screening method that could detect early stage colorectal neoplasia would be ground breaking (32, 33). CRC is initially presented as benign, polyp, the most aggressive of which become malignant, promoting metastasis to lymph node and secondary sites. Although not all adenomas progress to cancer, these are marked different in disease progression between patients. Progression from adenoma to CRC is driven by the loss of functional mutations in tumor suppressor genes such as APC and TP53 and also due to activation of mutations in oncogenes including Kristen RAS (KRAS). Mutations in KRAS promote cell growth by prolonging the activation of RAS/MAPK regulated signaling pathway which suggests that increase in mutation is negatively associated with survival. miR-224 is one of the several miRNAs, found to target KRAS. Detailed analysis of which revealed that it alters cell proliferation, invasion and Epithelial Mesenchymal Transition (EMT) phenotypes. It is also clearly evident that miR-224 is differentially expressed in colorectal adenomas and cancer which is an indication of an early event that occurs during the development from adenomas to cancer. Due to its significant importance in KRAS regulation, miR-224 is proposed to be used clinically as a marker that indicates disease progression (34).
Numerous studies showed that miRNAs play central role in various pathological and physiological processes. The pathogenesis of CRC depends on several signaling pathways including the TP53, PI3K, RAS, MAPK, EMT transcription factors and Wnt/β catenin pathways. And, CRC also depends on a plethora of miRNAs target inflammatory signaling molecules to induce or inhibit chronic inflammation and inflammation-related cancers. An investigation indicates DICER and PTEN are the target genes of miR103 and the up regulation of miR-103 represses cell proliferation and migration. In order to confirm in vitro results, xenograft models were generated by HCT116 cells with up regulated or down regulated miR-103 levels, in which up regulation of miR-103 significantly promoted cell proliferation and vice versa. Collectively, both in vitro and in vivo results showed miR-103 may act as a potential target for CRC via DICER and PTEN regulation (35). Transglutaminase-2 (TG2) activity has been linked to different biological processes associated with tumor development and progression, such as cell adhesion, motility, invasion, apoptosis, chemoresistance and epithelial-mesenchymal transition. However, some published studies demonstrated its contradictory functional roles such as promoting or inhibiting apoptosis. This behavior of TG2 may be related to cellular location and the availability of it’s identified many protein substrates, or to the balance between different isotypes of the enzymes. Cellura and his coworkers, with the aid of in silico analysis, identified that TG2 is targeted by miR-19, the fact was confirmed experimentally too. Over expression of miR-19 lead to subsequent decrease in TG2 expression which links to amplification events of chromosome13, leading to altered invasive behavior of CRC cells. Further investigation on this aspect may provide the potential to provide useful insights into the mechanisms driving the disease progression (36, Figure 3).
 Figure 3
Figure 3Pathways of miRNA induced colorectal cancer. Down regulation of tumor suppressor genes (APC, TP53) induce K-RAS mutation that prompts MAPK signaling. The cancer initiation and progression are then mediated by several miRNAs out of which miRNA 103 is clinically targeted.
Growing evidences prove that miRNA regulates different target genes by playing opposite roles as tumor suppressor in some cancer types but as oncogenes in others, whose detailed study is essential to elucidate its role in tumorigenesis. miR-126 is one such miRNA, located within intron 7 of the epidermal growth factor-like domain 7 gene (EGFL7), promote embryonic angiogenesis by promoting vascular endothelial growth factor (VEGF) signaling in zebrafish and in mouse model. miR-126 is found to be down regulated in breast and pancreatic cells while alleviated expression of miR-126 in non-small cell lung cancer and renal cell carcinoma was significantly correlated with the reduced patient survival. Although targeting oncogenic genes, such as SLC7A5, Crk and ADAM9, miR-126 also plays a role as a tumor suppressor by inhibiting tumor cell proliferation, invasion and the EMT process. In contradiction to this, miR-126 over expression contributes to gastric carcinogenesis by inhibiting SOX2 expression, and is highly associated with metastasis of prostate cancer. These contradictory results imply that miR-126 may function by diverse mechanisms in different physiological and pathological contexts (37).
DNA methylation and associated silencing of tumor-suppressor genes are few molecular hallmarks of human tumors. Recently, this phenomenon has been extended to miRNAs with tumor-suppressor features, which are down regulated in multiple tumors. Zhang et al. observed that extensive promoter methylation occurs in 52 out of 62 primary CRC tissues and 6 cell lines. In addition, treatment of the CRC cell lines with 5-aza-CdR, an inhibitor of DNA methyltransferase, restored miR-126 expression and thereby leads to a decrease in VEGF expression. It suggests that silencing of miR-126 by promoter methylation is an important mechanism underlying deregulation of VEGF expression in CRC (37). miRNAs could serve as potential biomarkers in CRC diagnosis based on its high degree of stability, specificity and sensitivity. The roles of miRNAs in tumor growth and the regulation of tumor progress summarized here suggest miRNAs could be a potential means for diagnosis and treatment as well as prognostic parameters for CRC. Ultimately, the goal is to identify clinically relevant miRNA biomarkers that are cost effective, and are easily assessed in a clinical setting, and contribute to patient management decisions, resulting in direct benefits of the patient.
MicroRNAs are believed to control the activity of more than 50% of all protein-coding genes. miRNAs are expressed in a tissue specific manner, thereby greatly contributing to cell-type-specific profiles of protein expression. It has been noted that several miRNAs drive lung cancer (Figure 4). Mainly, out of 30 observed miRNAs, 23 miRNAs are found to be highly expressed in lung of both mice and human. Out of 23, five miRNAs such as miR-26, miR-29, miR-30, let-7 and miR-99 are found to be highly expressed in common (38).
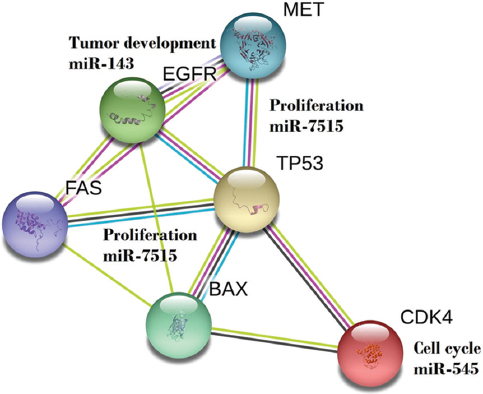 Figure 4
Figure 4Involvement of miRNA in pathways of lungs cancer. For proliferation, both miRNA 143 and 7515 are responsible, where as for tumor development miR-143 is responsible. Due to Mesenchymal Epithelial Transition (MET), tumor develops and/or proliferate in lungs. Role of Epidermal growth factor receptor (EGFR) and Tumor protein p53 and other cited molecules are associated with the event of transition.
Varied expression profiles of miRNAs proved that it can also be detected in plasma or serum, and their stability in serum can serve them as new potential biomarkers for diagnosis and prognosis. Recent study by Fortunato et al. substantially proved that the circulating miRNA levels are associated with prognosis. Microarray miRNA profiling of tumor tissues collected from 19 lung cancer patients indicates that eleven were mature miRNAs and were from the tissue and six miRNA precursors were found to be from the plasma of patients. Further, pathway enrichment analysis of these miRNAs confirmed their involvement in relevant biological processes, such as proliferation, growth factor receptor signaling, signal transduction, cell survival, dissemination, and metastasis; features that are altered in the more common cancers, and particularly in lung cancer (39).
Emerging data proves that miRNA scores an important in the mechanism-based therapeutic targets in cancer. Unique miRNA signature pattern may serve as molecular biomarkers for tumor diagnosis, classification, prognosis of disease specific outcomes and prediction of therapeutic response. miR-545 is less abundant in human lung cancer tissues than in adjacent non-cancerous lung tissues, and inhibits the proliferation of lung cancer. It also induces cell cycle arrest at G0/G1 phase and triggers apoptosis in lung cancer cells both in vitro and in vivo by repressing the expression of cyclin D1 and CDK4 genes, suggesting its functions as a tumor suppressor in lung cancer (40). miR-663, miR-365 and miR-7515 targets TFGβ1, NKX2-1 and human mesenchymal-epithelial transition factor (c-Met), respectively, thereby control over cell proliferation, differentiation, migration and apoptosis is gained (41, 42, 43). An enhanced study on these miRNAs and their deregulation in lung cancer cells may contribute in better understanding of the molecular mechanisms responsible for lung cancer.
Lung cancer incidence rates are approximately twice as high in developed countries (61/100,000 males and 19/100,000 females) compared with developing countries (29/100,000 males and 10/100,000 females). Non-small cell lung cancer (NSCLC) accounts for ~80% of all lung cancer cases. Despite recent advances in the diagnosis and chemotherapeutic and targeted treatment of NSCLC, the overall survival rate of NSCLC patients remains low (5-year survival rate of 15%) and the recurrence rate of NSCLC remains high, even with early diagnosis. In NSCLC, the leading cause of death is chemotherapy resistance and metastasis. As known, lung cancer arises from a series of genetic and epigenetic alterations that inactivate tumor suppressor genes and activate oncogenes. However, the basic mechanisms underlying lung cancer initiation and progression remains largely unknown. A greater understanding of the molecular mechanisms underlying carcinogenesis, progression and drug resistance in lung cancer would be helpful in improving diagnosis and prevention. Various miRs, including miR-183, miR-21 and miR-155, have been shown to promote the development of NSCLC. Conversely, miR-99a, miR-340 and miR-223, were reported to function as tumor suppressors in NSCLC. miR-143 was demonstrated to up regulate the mRNA expression levels of melanoma 2 and apoptosis-associated speck-like protein containing a CARD in a Jurkat cell line. Over expression of miR-143 reduces the survival rate of a cecal ligation and puncture-induced sepsis in mouse model. However, the function of miR-143 in NSCLC remains poorly understood. Furthermore, miR-143 was down regulated in NSCLC patient tissues and NSCLC cell lines. The over expression of miR-143 in NSCLC cells were able to suppress tumor growth, potentially by inhibiting EGFR expression. miR-143 in NSCLC, regulate the processes of cancer cell migration, invasion, proliferation and apoptosis (44).
As paclitaxel and cisplatin based chemotherapy is still the main treatment option for advanced metastatic lung cancer, chemotherapy resistance frequently drives tumor progression. Shen et al. identified that miRNA has vital role in chemosensitivity which might be helpful in overcoming drug resistance (45). It was observed that miR-137 was down-regulated in the human lung cancer tissues and in the resistant cells strains such as A549/paclitaxel (A549/PTX) and A549/cisplatin (A549/ CDDP) when compared with lung cancer A549 cells. Moreover, the over expression of miR-137 inhibits cell proliferation, migration, cell survival and cell cycle arrest in G1 phase in A549/PTX and A549/CDDP cell lines. Furthermore, repression of miR- 137 significantly promoted cell growth, migration, cell survival and cell cycle G1/S transition in A549 cells. The tumor suppressive role of miR-137 was mediated by negative regulation of Nuclear casein kinase and cyclin-dependent kinase substrate1 (NUCKS1) protein expression. This clearly indicates that miR-137 plays a prominent role in tumor progression which results in chemosensitivity in lung cancer. Now despite having a better understanding of the molecular events that govern the lung cancer than ever before, it remains a clinical challenge in the treatment of lung cancer and these miRNAs may offer a new modulation strategy to overcome chemoresistance (45).
Pancreatic cancer (PC) is one of the most fatal malignancies with increasing incidence and high mortality all around the world. Less than 10% PC patients are diagnosed at an early stage, and most patients do not have opportunity of surgical resection due to relatively late stage. Despite recent advances and efforts in the treatment, the 5-year overall survival rate of PC is lower than 5%. This discouraging fact reflects the lack of improvement in detection and diagnosis strategies and the paucity of breakthroughs in treatment regimens. Current investigations are much more focused on miRNA which are very well known to participate in the development and progression of cancer (46, 47, 48). A decade before, Lee et al. studied the miRNA signature pattern in pancreatic cancer. They performed miRNome profiling of 200 miRNAs, in specimens including human pancreatic adenocarcinoma, paired benign tissue, normal pancreas, and pancreatitis cell lines. Among them, about 100 miRNAs are expressed aberrantly, but only four miRNAs (miR-155, miR-21, miR-221 and miR-222) are found to be expressed commonly in other cancer types and specifically miR-376a and miR-301 seemed to be confined to pancreatic cancer. They noted that miR-221, miR-21 and miR-301 are the prominent miRNA that are differentially expressed and are found to be localized in tumor cells and not in stroma or normal acini or ducts (49). A total of 167 PC patients were enrolled for the study of miRNAs associated with overall survival in PC patients by Cox proportional regression model. Thirteen miRNAs were identified to be significantly related with overall survival in PC patients. Patients with high risk score suffered poor overall survival compared with patients who had low risk score. The multivariate Cox regression analyses showed that the miRNA signature could act as an independent prognostic indicator. Their study identified a miRNA signature including 13 miRNAs which could serve as an independent marker in prognosis of PC. Zhou et al. concluded from their findings that by analyzing the genome-wide miRNA expression profiles which could act as an indicator for treatment outcome and could be served as an independent factor in prognosis of PC (47, Figure 5).
 Figure 5
Figure 5One of the pathways in pancreatic cancer initiated by miRNA 141. Involvement of other miRNA such as 21, 221 301, 376 are also involved in developing pancreatic cancer.
Precision medicine is a treatment targeted on the basis of various factors such as genetic, biomarker, phenotypic, or psychosocial characteristics to particular individual. In the context of cancer treatment, technology advances and increase in molecular understanding of the etiology of pancreatic cancer have allowed the precision medicine to impact patients. Discovery of common genetic mutations that drive different cancers (such as KRAS) has facilitated the development of more individualized drugs targeting relevant mutations found in each tumor type. For the past few years, miRNAs are explored for their regulation of hundreds of cancer related genes and pathways that drive cancer (50). For instance, miR-141 is down regulated in tissues and cell lines of PC and found to be in correlation with tumor size and TNM stage as well as lymph node and metastasis. Using bioinformatics tools, MAP4K4 is predicted as its target which is later validated by dual luciferase reporter gene assay. Furthermore, knockdown of MAP4K4 was found to inhibit cell proliferation, clonogenecity and invasion, induced G1 arrest and apoptosis and was associated with the enhanced chemosensitivity. Additionally, over expression of miR-141 was observed in a nude mouse xenograft model and knockdown of MAP4K4 was significantly found to suppress PC cell growth. MAP4K4 belongs to the mammalian STE20/MAP4K family, associated with cell motility, rearrangement of the cytoskeleton, and cell growth. Regulation of MAP4K4 by miRNA proves its critical role in cancer cell proliferation, cell cycle, and apoptosis (51). Deng et al. noted that miR-25 had significant diagnostic information in comparison with serum tumor marker carcino embryonic antigen (CEA) suggesting serum miR-25 has strong potential as a novel biomarker for the early detection of PC (52). These data provide clear insights into the miRNA-driven pathophysiological mechanisms. It offers new candidate targets to be exploited both for diagnosis and for the application of precision medicine. It can minimize unnecessary side effects present in broad, traditional treatment, improve clinical outcomes, reduce rates of toxicity, and antibiotic resistance.
Gastric cancer (GC) is the fourth most common cancer in the world, and has the second highest cancer-related mortality rate. Although more progress has been achieved in recent years, the early diagnosis and treatment for gastric cancer are not yet satisfactory (53). miRNA expression could be used as a novel strategy for chemoprevention of human gastrointestinal cancer because miRNAs have been found to regulate a variety of cellular processes such as cell proliferation, differentiation, invasion, migration, and EMT. In detail, miRNAs negatively regulate the expression of cancer-related genes by decreasing the expression of tumor suppressor genes or enhancing the expression of oncogenes, or as modulators of cancer stem cells and metastases (54). For instance, miR-331-3p is down regulated in GC which clearly indicates its role in enhancing cell proliferation. Restoration of miR-331-3p blocked G1/S transition on SGC-7901 and AGS cell lines by interfering E2F1 activity (55). Similarly, Yin et al. showed decrease in miR-205 levels promoted cell proliferation by up regulating YinYang1 expression, oncoprotein via targeting its 3’ UTR (56). In contrast, miR-106a is up regulated in GC. Assessment of Caspase-8, PARP and Caspase-3 has led to the revelation that miR-106a represses the cell growth by triggering apoptosis by interfering with Fas activity (57). In another study, the ectopic expression of miR-433 and miR-127 was found to suppress the cell proliferation, progression, migration and invasion by targeting KRAS and MAPK4 oncogene in HGC-27 gastric cancer cell line (58). Accumulating evidence suggests that miRNAs play an important role in GC, but the role of specific miRNAs involved in this disease remains elusive.
miRNA expression profiling may be used as a powerful clinical tool for cancer diagnosis. To study the role of miRNAs in gastric cancer, Yao et al. studied the expression profile of 847 miRNAs in Chinese patients with GC. A total of 24 miRNAs with more than 2-fold change were differentially expressed between normal and cancerous gastric tissues. Of these, 22 miRNAs were found to be significantly up-regulated in GC, whereas only miR-638 and miR-378 were significantly down-regulated in GC compared to normal gastric tissue. It suggests the involvement of these miRNAs in the development and progression of GC (59). In another study, among these 38 miRNAs, miR-34b, miR-127-3p, miR-129-3p and miR-409 were found adjacent to CpG islands. The methylation-silenced expression of these miRNAs could be reactivated in GC cells by treatment with demethylating drugs in a time-dependent manner. Analysis of the methylation status of these miRNAs showed that the upstream CpG-rich regions of miR-34b and miR-129-2 are frequently methylated in GC tissues compared to adjacent normal tissues, and their methylation status correlated inversely with their expression patterns. The expression of miR-34b and miR-129-3p was down regulated by DNA hypermethylation in primary gastric cancers, and the low expression was associated with poor clinicopathological features (60, Figure 6).
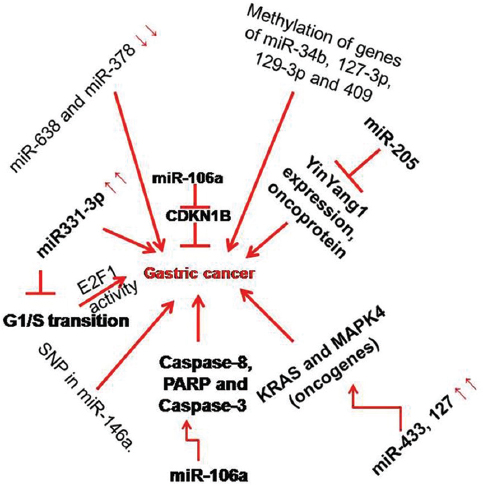 Figure 6
Figure 6Pathways of gastric cancer and their regulation by miRNA. Caspase3, 8 and CDKN1B intermediates are associated with specific miRNA as indicated in the figure. Processes such as SNP in miRNA-146a also induces gastric cancer.
Gastric cancer can be caused by personal genetic susceptibility, lifestyle factors, and environmental factors. However, some researchers have found that only a limited population exposed to these risk factors ultimately develops gastric cancer. Considering this fact, genetic susceptibility may play a significant role in the development of gastric cancer. Single-nucleotide polymorphisms (SNPs), a major type of genetic variant, have been widely investigated as molecular markers to predict the initiation, treatment outcomes, and prognosis of gastric cancer. Recently, miRNA SNPs have been reported in several studies. One of the most described miRNA SNPs associated with elevated risk in GC is SNP rs2910164 of miR-146a. Ahn et al. (61) demonstrated that the C/G polymorphism in miR-146a decreases miR-146a expression and subsequently leads to the reduced regulation of the target genes TRAF6, IRAK1 and PTC1 by the C allele. Thus, SNP rs2910164 may be used as a genetic biomarker to predict GC risk. SNPs in pri-miRNAs and pre-miRNAs could affect the maturation process and function of the miRNA, which may affect the expression of many proteins in the signaling pathway (61). Up regulation of pri-let-7a-2 expression by the rs629367 C/C genotype was associated with increased risk and low survival in GC, probably by affecting the expression of mature let-7a. The binding capacity of a miRNA with its target can be modified by SNPs affecting the miRNA TAG sequence. Additionally, a SNP in mRNA sequence could influence the complementarity between the miRNA and the target mRNA. This could result in alteration of susceptibility to tumorigenesis. Wang et al. described that a SNP in the PDL1 (rs4143815) could affect its protein expression by interfering with miR-570 negative regulation (62). Furthermore, this SNP was significantly related to the risk of GC and depth of tumor infiltration, differentiation grade, lymph node metastasis, tumor size and staging. Hence, SNP data could be useful to improve our understanding of the contribution of individual susceptibility to GC pathogenesis (63, 53).
Acute lymphoblastic leukemia (ALL) is the most common hematologic malignancy in children and its occurrence is noted in 2 to 5 old children. French-American-British cooperative group classification of ALL had been deserted because it failed to meet clinical relevance. Later, WHO developed the classification based on morphology, immunology, cytogenetics and molecular biology while immunophenotyping is based on cell surface and cytoplasmic proteins. And, it is currently being practiced. According to immunophenotyping, ALL is classified into two types, T-ALL and B-ALL. The important markers of T-ALL includes the terminal deoxynucleotidyl transferase (TdT), CD2+, CD3+, CD4+, CD5+, CD7+ and CD8+. B-ALL is further subdivided into early pre-B-cell, pre-B-cell and mature B cell. The important markers for the subtypes are as follows: early pre-B-cell includes TdT+, HLA-DR+, CD19+, CD34+ and CD10+; pre-B-cell includes TdT+, HLA-DR+, CD19+, CD10+ and CD20+ and the markers of mature B-cell are HLA-DR+, CD19+, CD10+, CD20+ and surface Ig (sIg)+. In addition to the immunophenotyping of ALL, large number of studies showed that the miRNA expression profiles have strong impact on the development of leukemia (64).
The basic helix-loop-helix transcription factor TAL1 is necessary for hematopoietic commitment which is expressed very early in the hematopoietic differentiation but down regulated during the early stage development of thymic progenitors in T-cell lymphopoiesis. About 60% of T-ALL patients found to express the increased TAL1 transcripts. The microdeletion of TAL1 locus is the most frequent chromosomal alteration which fuses the TAL1 coding region to the SLL regulatory elements, producing the SIL-TAL1 fusion gene. This modification is found to occur in 9-25% of childhood T-ALLs. The mechanism of aberrant activation of TAL1 in T-ALL patients is largely unknown. Correia et al. showed that TAL1 levels are regulated partly by miRNAs (65). They performed computational prediction of miRNAs that bind to the TAL1 mRNA in which miR-101, miR-520d-5p, miR-140-5p, miR-448 and miR-485-5p are found to be possible miRNA candidates. Further experiments showed that over expression of these miRNAs in different T-ALL cell lines resulted in the consistent down regulation of TAL1 protein. In coherence, inhibition of miR-105 and miR-520d-5p promoted TAL1 protein expression. They also found that miR-101, miR-140-5p, miR-448 and miR-4855p were down regulated in T-ALL patient samples as well as in T-ALL cell lines. This results show the existence of relation between the TAL1 and specific miRNAs which contributes to the ectopic expression of TAL1 in T-ALL (65).
MLL-AF4 (mixed-lineage leukemia- AF4 fusion protein) plays a significant role in leukemic clonogenicity and also attributes to the aggressive nature of ALL. The chromosomal translocation of MLL is seen nearly 5% in ALL and accounts for over 70% cases of infant leukemia. More than 50 leukemogenic fusion proteins are coded by MLL translocations. The most frequent is t(4:11)(q21;q23) which encodes the MLL-AF4 fusion protein. ALL patients with MLL-AF4 shown to have high relapse rate and poor prognosis. Previous studies demonstrated that over expression of MLL-AF4 fusion gene induces resistance to etoposide mediated cytotoxicity whereas, the reduced expression of MLL-AF4 induces apoptosis and also controls the proliferation of tumor cells (66). In 2009, Kotani and his coworkers found that MLL-AF4 was down regulated by miR128b while its downstream transcriptional target cyclin-dependent kinase inhibitor 1B (CDKN1B) was suppressed by miR-221. Sequential study by Kotani in 2010 revealed that glucocorticoid resistance was related to downregulation of miR-1286 or miR-221 and the restoration of miRNA levels was therefore proposed as a promising therapy for MLLAF4+ in ALL (67, 68). With this background Duo et al. focused on MLL-AF4 and miRNAs. They identified MLL-AF4 as direct target of miR-143 and on restoration of which was found to induce apoptosis in MLL-AF4 in ALL leukemic cells. However, they focused on miR-142-3p because it was found to be a better target for MLL-AF4. The results showed that upregulation of miR-142-3p was able to decrease in MLL-AF4 expression in the RS4;11 leukemic cell line (Figure 7). Ectopic expression of miR-142-3p remarkably suppressed cell proliferation and also induced apoptosis in RS4;11 cells expressing the MLL-AF4 fusion protein. The exogenous expression of miR-142-3p was to highly reduce the expression of MLL-AF4 target genes such as homeo-box A (HOXA) 9, HOXA7, and HOXA10 (66).
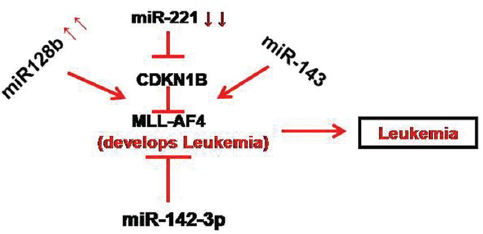 Figure 7
Figure 7Regulation of leukemia by several miRNA. Most of the miRNA are seem to be associated with mixed-lineage leukemia- AF4 fusion protein (MLL-AF4), a molecule associated with aggressive leukemia.
Leukemia is a heterogenous disease which is characterized by the abnormal proliferation of blood cells of myeloid or lymphoid origin. Till now, numbers of miRNA are found to be involved in leukemia. Through their findings, Zhu et al. supported that miR-128, miR-223, miR-181a and let-7 may have diagnostic value in ALL while miR-181a and miR-155 are of great prognostic significance in AML (69). Further, detailed analysis of miR-181a was carried out by Dahlhaus and coworkers who demonstrated that high mobility Gp-protein B1 (HMGB1) and cluster of differentiation (CD4) would be the possible target of miR-18a. They also found that increase in expression of miR-181a lead to the suppression of cell proliferation and metabolic activity in T and B cells in ALL as well as in AML (70).
The first evidence of a correlation between miRNAs and cancer was reported by Calin et al. (71, 72) who observed knockdown or knockout of miR-15a and miR-16-1 in approximately 69% of Chronic Lymphocytic Leukemia (CLL) patients. Immediately after these initial observations, all the known microRNA genes were mapped and it was found that they are frequently located in cancer associated genomic regions (CAGRs). These regions usually include minimal regions of amplification, loss of heterozygosity (LOH), common breakpoint regions in or near oncogenes (OGs) and tumor suppressor genes (TSGs), and fragile sites (preferential site of chromatid exchange, deletion, translocation, amplification, or integration of plasmid DNA and tumor-associated viruses) (71, 72). Teichler et al. described the negative correlation of miR-29a with its target SK1, a nuclear oncogene (73) whereas Garzon et al. reported on miR-29b which targets DNMT 3a and 3b directly and also indirectly targets DNMT 1 through Sp1, a transactivator of the DNMT 1 gene indirectly (74). miR-15a and miR-16-1 are repressed by inversely correlating Bcl2, antiapoptotic protein in CLL (75). Another miRNA expression profiling was assessed in cell lines, patient samples of CML in comparison with normal cells. Rohak and coworkers observed 3 miRNAs miR-31, miR-155, miR-564 that are downregulated while Agirre et al. noted that miR-10a and miR-150 along with other mentioned miRNAs are downregulated while miR-96 alone upregulated in CML (76, 77). Data of Xu et al. suggest that miR-138 is found to be downregulated in CML. BCR-ABL regulates miR-138 expression by inhibiting the expression and transcriptional activity of GATA1. Addition of imatinib inhibited BCR-ABL activity and activates GATA1, which upregulates miR-138 transcription. In conclusion, they have identified a novel BCR-ABL/GATA1/miR-138 regulatory circuitry whose dysfunction may contribute to the pathogenesis of CML (78). It is known that altered miRNA expression play a vital role in the pathology of leukemia, thus miRNAs have rapidly emerged as potential targets for therapeutics. Recent studies showed the important roles of miRNAs in the pathogenesis of leukemia, including AML, ALL, chronic myeloid leukemia (CML) and CLL (79). As a consequence of their ability to regulate gene expression, microRNAs are involved in the most crucial cellular processes, spanning from development, differentiation, cell cycle regulation to senescence and metabolism, and their expression is aberrant in several human diseases, including cancer.
Aging is considered as increase in metabolic dysfunctions in cells mostly linked to adverse structural and functional changes at organ level. Such events lead to a progressive loss in the efficiency of different physiological functions that may consequence as increase in susceptibility to disease, finally, leading to death (80). The “free radical theory of aging” is one of the most believable and acceptable explanations for the mechanistic basis of aging. It postulates that aging and its related diseases are the consequence of free radical-induced damage to cellular macromolecules and the inability to counterbalance the produced high level of active oxygen molecules by endogenous anti-oxidant defenses. Therefore, ageing has a strong positive correlation with phenomena such as oxidative damage, structural modifications and loss of potency of several biomolecules including enzymes in animals (81, 82). The “rate of living” is accepted as aging and according to modern aging theory, the “rate of living” is defined by metabolic dysfunction rather than the loss of vital substance. However, advances in aging research in 21st century continue to challenge this theory at several checkpoints. The debate in the fiction still exists to accept whether metabolic rate or metabolic stability is the determinant factor for longevity. Therefore, nine candidates such as genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion and altered intercellular communication are proposed to be the cellular and molecular hallmarks of aging (83).
Many of the above factors are directly or indirectly found to be the “cause” or “effects” of ageing in animals (84). Recently, Missios et al. reported the first experimental evidence that telomere dysfunction enhances the requirement of glucose substitution for the maintenance of energy homeostasis and IGF-1/mTOR-dependent mitochondrial biogenesis in progressively aging tissues (85). Indeed, the mitochondrial rate of free radical production seems to have a much stronger correlation with maximum longevity in animals (86, 87). On the other hand, age associated markers found to have strong correlation with cancer and its associated complications (88). Aged body increases a greatest risk factor for developing cancer. A statistics indicates that 60% of people at a specific time point who have contracted cancer are of about 65 years or older. Even a study reported that the majority of patients with cancer in the United States are more than 70 years old (89). So, it indicates age is one of the important factor in developing cancer. The chance factor is enhanced due to elderly complications such as high blood pressure, heart diseases, lung disease, diabetes, kidney diseases and arthritis disorders in elderly people (90).
Finkel et al. reported that “at first glance, cancer and ageing would seem to be unlikely bedfellows (88). Therefore, it is argued that yet the origins for this improbable union can actually be traced back to a sequence of tragic-and some say unethical-events that unfolded more than half a century ago.” Thereafter, a series of observations tried to impose a complex but growing convergence between the biology of ageing and the mechanisms that underlie cancer (91-99). Over the last 60 years, major improvements in our understanding on the driving forces for aging and cancer, whether parallel and opposing, has become clearer. Mechanism of telomere shortening, senescence, and adult stem-cell regulation in aged animals gained support from cumulative experimental evidences and proved a positive but not limited correlation between cancer and aging. Accumulation of mutations being the common factor for aging and cancer, and, down regulation of tumor suppressor pathways with misbalance in maintaining energy homeostasis in aged tissues contribute to cancer. According to hyper-function theory, aging is a quasi-program favoring both age-related diseases and cancer that could be inhibited by the regulation of longevity pathways and therefore, both are supposed to have a positive correlation (100). So, it is concluded that the incidence of cancer increases with age in both humans and laboratory animals. Thereafter, it was proposed that age-related increase in cancer incidence in human and non-human laboratory model animals would follow the same mechanism (101). As a result, contrasting patterns of age-related distribution of tumors in different organs and tissues are observed in various studies. Aging may increase or decrease the susceptibility of various tissues for the initiation of carcinogenesis but it is confirmed that it usually facilitates promotion and progression of carcinogenesis. The latter case follows several mechanisms. Tissue accumulation of cells in late stages of carcinogenesis with alterations in internal homeostasis including change in immunity and endocrine secretions. Increase in expression of tumor promoters is found to be a common cause in both aged animals and aged humans. The reasons are attributed to several age associated molecular, cellular and physiologic adverse events that influence carcinogenesis and subsequent cancer growth (101). Therefore, it is elegantly proved that aging drives both degenerative and hyper-plastic pathologies, most likely by promoting chronic inflammation and cancerous growth in human and non-human laboratory models (102).
Since a number of studies have correlated the role of miRNA with cancer, now it is pertinent that an intricate relation must exist among cancer, aging and expression of miRNA. Genes that controls major cellular events such as changes in DNA repair and DNA damage response, telomere shortening, changes in control over the expression and regulation of genes brought about by change in epigenetic and mRNA processing, loss of protein homeostasis, altered nutrient signaling, mitochondrial dysfunction, stem cell exhaustion, premature cellular senescence and altered intracellular communication are also controlled by miRNA (103). The above events are considered as the hallmarks of aging. Therefore, degeneration of organismal, tissue and cellular homeostasis related to miRNA regulation are considered as potential biomarkers for healthy ageing. MiRNAs have pivotal roles in post-transcriptional regulation of gene expression. Table 2 gives a clear idea about the particular miRNA regulating different cellular events associated with aging.

|
Processes such as DNA methylation induced silencing of miR-124a, miR-34, miR-9 and miR-200 gene families indicate their regulation pathway. MiRNAs miR-15a, miR-16 and miR-29 have been shown to demonstrate lower expression upon increase in expression of HDAC1, HDAC2 and HDAC3 transcripts, which are responsible for histone modifications and it may therefore linked to telomere shortening. Aged cells are also immune-compromised and prone to inflammation and both the two major processes are associated with the action of miRNA (104). MiRNA molecules discharge a complex and indirect role in the process of protein synthesis both qualitatively and quantitatively from the genetic makeup. Since protein production analogously changes with respect to aging, it is argued that the miRNA system must react to the cellular and molecular damage that causes aging, as another part of the reaction (105-108). Therefore, there is every reason to believe that miRNAs play a major role in modulating life span and the aging process; indeed, support for this assumption has emerged from several studies on wide variety of model organisms. For example, miRNA lin-4 (a longevity promoting factor) and its target lin-14 (a life span antagonizing factor) control lifespan post-developmentally in C. elegans (105, 106, 107, 108). Similarly mir-71, -238 and -246 mutants display a significantly shorter lifespan than those of wild type animals, and over expressing miR-71 or miR-246 increases lifespan, indicating that these miRNAs function to promote longevity (104). Nearly 200 miRNAs, for example miR-34, in C. elegans have shown altered expression in different stages of life and >50% of them have conserved sequences in humans (108-110). So, it can be predicted that miRNA has a definite role in aging of human and non-human animals (108, 109). Genetic studies have demonstrated that the lin-4 miRNA and its target lin-14 function in the same pathway as DAF-2 and DAF-16, two conserved genes of aging pathways in C. elegans. And, miR-1, miR-320, and miR-206 targets IGF-1, and miR-216a, miR-217, and miR-21 targets PTEN which are related to genes of aging pathways (111- 114). It is also recorded that age-associated diseases are also regulated by miRNA. Insulin/IGF signaling, DAF-12 signaling and TOR signaling that are related to several human diseases including heart, muscle and neurodegenerative disease are targeted by miRNAs such as miR-1, miR-21, miR-122 and miR-375 (115-118).
Aging and its associated consequences including cancer under normal and varied environments are now considered as few of the important areas of research, not only in human beings but also in other non-human model organisms (119-123). Although study on the synergistic role(s) of miRNA on aging and cancer is scanty, many of them are found to be associated with the events that control both the above processes (124-126). For, example DNA damage is a genomic event that tremendously affects both aging and cancer. Many miRNA such as miRNA 21, 102, 374 and 504 are believed to dramatically influence metabolic pathways related to DNA damage. Micro RNA 1, 301b, 320 etc. shape the fate of protein homeostasis, a major factor that lead to aging and cancer. Inflammation related pathways that are also possibly influence both the events in cells and are also regulated by miRNA such as 29C, 290, 293 and 499. Few miRNA such as miRNA190b are found to be used as biomarkers of breast cancer and nutritional sensing (an event closely associated to aging). MicroRNA137 and 143 control gene regulation (regulates aging) and lung cancer. Gene regulation and gastric cancer are synergistically influenced by miRNA 127. Finally, gastric cancer and cellular senescence are regulated by miRNA 106a strongly confirms that miRNA are closely associated to regulate the molecular events in both aging and cancer. Therefore, targeting these common miRNA in future may be helpful to develop therapeutic approaches to minimize the up regulation of molecular pathways that influence aging and cancer (127-130) (Table 1, 2 and Figure 8).
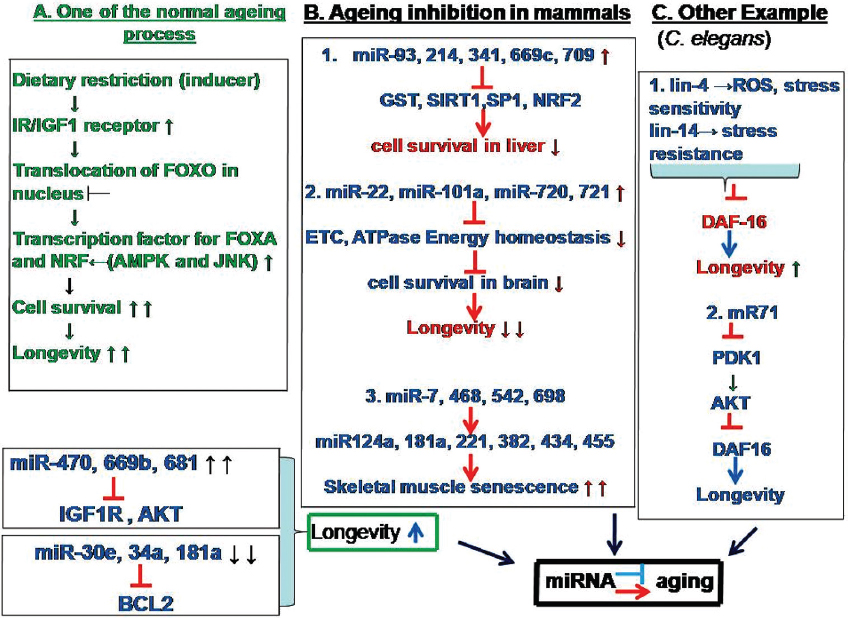 Figure 8
Figure 8Pathways for the role of miRNA in aging. A. Out of many ageing processes, transcription of FOXO down regulation in nucleus mediate slowed aging in which IGF1R (Insulin-like growth factor 1 receptor) and AKT (Protein kinase B (PKB) and BCL2 (B-cell lymphoma 2) are involved. B. Various processes that inhibit ageing in mammals by different miRNA in different tissues by influencing at electron transport chain (ETC) level, enzymes like glutathione-S-transferase (GST), nuclear factor (erythroid-derived 2)-like 2 (NRF2), and Sirtuin 1 (SIRT1). C. Example of the involvement of miRNA in aging in C. elegans. Genes such as DAF family and AKT may mediate reactive oxygen species (ROS) to increase longevity. Therefore, an intricate relation exists between aging and miRNA.
microRNAs (miRNAs) are evolutionarily conserved, small, noncoding RNAs of about 22 nucleotides in length that function as posttranscriptional gene regulators. This review summarized the key mechanism of miRNAs and their regulatory functions towards specific cancers including their role in cell proliferation, differentiation, metastasis, and angiogenesis and as onco-miRNAs and tumor suppressors. Data regarding the changes in miRNA expression analyzed in this article provide insights into comprehensive mechanism of expression of miRNAs by genome wide expression, prediction of their target based on bioinformatics tools and in vitro and in vivo assays in varied cancers. Despite the current knowledge, understanding the complexity of miRNAs and all the other factors involved in modulating their interaction with genome is of fundamental importance in health and disease. Since there is pressing need for clinical translation of novel breakthrough in cancer biology, increased attention revolves around miRNAs as novel gene regulators with potential to fine tune physiological process involved in cellular differentiation and metabolism including aging (Figure 9). Given that the deregulation of miRNA expression is very much implicated in different stages of cancer development and aging, miRNAs, as anticipated to be potential targets for diagnosis, prognosis and treatment of cancer and may be used as potential biomarker to detect rapid aging process. As a result, they also can be targeted to slower down aging process. However, in order to obtain the holygrail, more theoretical and experimental studies are warranted to clarify how the miRNAs regulatory network is controlled.
 Figure 9
Figure 9Regulation of aging and cancer associated events via of specific miRNA. The middle column indicates the miRNA that regulates the respective aging (left column) and cancer (right column) related events as indicated in the figure.
BRP is profoundly thankful to the Science and Engineering Research Board, Department of Science and Technology, Govt. of India New Delhi, India (No. ECR/2016/001984) and Department of Science and Technology, Government of Odisha (Grant letter number 1188/ST, Bhubaneswar, dated 01.03.17, ST-(Bio)-02/2017) for providing funding.
