- Academic Editor
Background: Lung cancer is one of the most serious malignant tumors
endangering human health and life. This study focused on evaluating the
association between single nucleotide polymorphisms (SNPs) of the glutaminase
(GLS) and lung cancer susceptibility in the Chinese Han population.
Methods: A total of 684 lung cancer patients and 684 healthy individuals
were enrolled. Five GLS SNPs (rs143584207 C/A, rs117985587 T/C,
rs74271715 G/T, rs2355570 G/A, and rs6713444 A/G) were screened as candidate
genetic loci. Odds ratios (ORs) and 95% confidence intervals
(95% CIs) were calculated to assess the association between GLS SNPs
and lung cancer susceptibility. False-positive report probability (FPRP) analysis
further verified whether the positive results deserved attention. Finally, the
multi-factor dimensionality reduction (MDR) method was applied to analyze the
interactions between SNPs. Results: The overall analysis revealed that
GLS rs143584207 and rs6713444 were significantly associated with lung
cancer susceptibility. The subgroup and clinical information analyses further
revealed that GLS rs143584207 and rs6713444 could remarkably reduce lung
cancer susceptibility in different subgroups (age
Lung cancer is one of the most common cancers and the main cause of cancer-related death in the world [1, 2]. The Global Cancer Statistics 2020 showed that the number of newly diagnosed lung cancer cases exceeded 2.2 million (about 11.4% of all newly diagnosed cancer cases), and the number of lung cancer deaths exceeded 1.8 million (about 18.0% of all cancer deaths) in the world [2]. Additionally, the morbidity and mortality of lung cancer ranked first among all malignant tumors in males, whereas, ranked third and second in females, respectively [2]. Lung cancer mainly includes two histological subtypes, namely small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), and NSCLC includes three pathologic subtypes: lung adenocarcinoma, squamous cell lung cancer, and large cell carcinoma [3, 4]. Numerous studies have revealed that lung cancer susceptibility is related to environmental factors (toxic workplaces, air pollution, smoking, etc.) and genetic factors (genetic mutation, gene polymorphisms, etc.) [5, 6, 7]. In particular, genetic factors have a vital role in the pathological mechanism of lung cancer [8]. So far, relevant studies have found that some genetic loci in certain genes are related to lung cancer susceptibility, for instance, vascular endothelial growth factor (VEGF), interleukin-32 (IL-32), and so on [9, 10, 11, 12, 13]. However, although the glutaminase (GLS) gene is closely correlated with the occurrence of various cancers, its relationship with lung cancer susceptibility has not been reported.
The GLS gene encodes a K-type mitochondrial glutaminase, which can catalyze the hydrolysis of glutamine to produce glutamate and ammonia. The GLS gene can be expressed in human cells as two subtypes, namely kidney-type glutaminase (KGA, also called GLS1) and liver-type glutaminase (LGA, also called GLS2) [14]. The expression of the GLS gene has an essential influence on the production of metabolic energy, the synthesis of the neurotransmitter glutamate in the brain and the maintenance of renal acid-base balance. Numerous studies have indicated that the aberrant expression of GLS, especially GLS1, plays a huge role in tumor metabolism and the development of various cancers, including hepatocellular carcinoma [15], breast cancer [16], colorectal cancer [17], intrahepatic cholangiocarcinoma [18], prostate cancer [19], lung cancer [20, 21], melanoma [22], and so on. Studies related to lung cancer have shown that the inhibition of tumor growth can be achieved by preventing or interfering with the metabolism of GLS in tumor cells. Momcilovic et al. [21] showed that the combined use of CB-839 and erlotinib in epidermal growth factor receptor (EGFR) mutant NSCLC may affect the utilization of glucose and glutamine by reducing the metabolism of GLS, and ultimately inhibit the growth of tumor cells. Galan-Cobo et al. [20] also found that the development of GLS inhibitors might be a good method in the treatment of KRAS mutant lung adenocarcinoma. The hydrolysis of glutamine promoted by the high expression of GLS is an important anaplerotic reaction for the proliferation and survival of many cancer cells, that is to say, cancer cells can replenish intermediates related to glutamine metabolism into the Krebs cycle, which not only provides Adenosine Triphosphate (ATP) for cancer cells but also provides precursors for the synthesis of macromolecules [23]. Relevant studies have further found that many oncogenes and tumor inhibitors are related to the expression of GLS and the regulation of glutamine metabolism, thus affecting the occurrence and development of cancers. For instance, selenite has been proven to inhibit the expression of GLS and the metabolism of glutamine, thereby inducing the dose-dependent apoptosis of a variety of cancer cells [23, 24]. Van den Heuvel et al. [14] have found that the instantaneous knockdown of GLS1 splice variant GAC can affect the decomposition of glutamine, thus adversely affecting the growth of lung cancer cells. In addition, relevant studies have demonstrated that gene polymorphisms may lead to changes in gene expression and the activity of cancer-related enzymes, thereby affecting the susceptibility to cancers [5, 25, 26]. Therefore, in this study, we speculated that GLS gene polymorphisms might be associated to the lung cancer susceptibility, and the specific mechanism might be that genetic variants of the GLS changed the activity of glutaminase and inhibited the decomposition of glutamine, thereby affecting lung cancer susceptibility.
In this study, five candidates GLS SNPs (rs143584207 C/A, rs117985587 T/C, rs74271715 G/T, rs2355570 G/A, and rs6713444 A/G) were successfully selected and genotyped. Among them, rs143584207 is a missense mutation, which may have a certain impact on the expression of GLS. The other four SNPs are located in the 3’-UTR region and do not change the amino acid sequence, but they may influence protein folding, and then, gene function, which may eventually affect the occurrence of diseases [27]. To preliminarily investigate the association between five candidate GLS SNPs and lung cancer susceptibility, we conducted a case-control study of 1368 Chinese Han subjects. The association between GLS SNPs and lung cancer susceptibility was assessed by the overall, subgroup and clinical information analyses, so as to provide a new idea for further probing into the complex pathogenesis of lung cancer and finding more effective treatments for lung cancer patients.
There were 684 lung cancer patients (207 females, 477 males) were enrolled from the Affiliated Hospital of Xizang Minzu University. These patients were diagnosed as primary lung cancer clinically and histopathologically at the early stage, and they had no history of other cancers, and no acute or chronic pathologies. Afterwards, with the help of the tumor, node, metastasis (TNM) staging system, we determined the clinical stages of patients [28]. And the pathological types of these patients mainly included lung adenocarcinoma and squamous cell lung cancer. In addition, 684 healthy individuals (193 females, 491 males) were recruited, and they underwent health examination annually and had no any personal or family history of malignant tumors, respiratory diseases, and endocrine or metabolic nutritional diseases. This study was conducted under the standards approved by the Biomedical Ethics Committee of Xizang Minzu University (No. 20200-11), and conformed to the ethical principles of the World Medical Association Declaration of Helsinki for medical research involving humans. Before participating in this study, all participants signed informed consent forms.
GLS SNPs were screened according to the detailed steps below: (1) The
location of GLS was determined in the e!GRCh37 database
(http://asia.ensembl.org/Homo_sapiens/Info/Index) and all mutation sites of this
gene were obtained; (2) Haploview v4.2 (Daly Lab, Cambridge, Massachusetts, USA) was applied to filter SNPs (parameters:
Hardy-Weinberg equilibrium (HWE)
G*Power v3.1.9.7 (Heinrich-Heine-Universität Düsseldorf, Dusseldorf, North Rhine-Westphalia, Germany) was used to calculate the sample size through independent
samples t-test. The parameters were set as: Tail = 2, Effect size =
0.20,
In the case group, 684 patients with lung cancer included 207 females (30.3%)
and 477 males (69.7%), and their average age was 60.18
| Characteristics | Case (n = 684) | Control (n = 684) | p | |
|---|---|---|---|---|
| Age (years) | Mean |
60.18 |
59.79 |
0.441 |
| 359 (52.5%) | 371 (54.2%) | |||
| 325 (47.5%) | 313 (45.8%) | |||
| Gender | Male | 477 (69.7%) | 491 (71.8%) | 0.405 |
| Female | 207 (30.3%) | 193 (28.2%) | ||
| Smoking | Yes | 407 (59.5%) | 399 (58.3%) | 0.660 |
| No | 277 (40.5%) | 285 (41.7%) | ||
| Drinking | Yes | 355 (51.9%) | 332 (48.5%) | 0.214 |
| No | 329 (48.1%) | 352 (51.5%) | ||
| BMI | 388 (56.7%) | 410 (59.9%) | 0.641 | |
| 296 (43.3%) | 274 (40.1%) | |||
| Tumor types | Adenocarcinoma | 314 (45.9%) | – | – |
| Squamous cell carcinoma | 219 (32.0%) | – | ||
| Stages | III–IV | 392 (57.3%) | – | – |
| I–II | 292 (42.7%) | – | ||
| Lymph node metastasis | Yes | 318 (46.5%) | – | – |
| No | 350 (51.2%) | – | ||
| Notes: | ||||
As shown in Table 2, five GLS SNPs (rs143584207 C/A, rs117985587 T/C,
rs74271715 G/T, rs2355570 G/A, and rs6713444 A/G) were screened and genotyped.
The functional prediction results revealed that the GLS SNPs were
associated with the promoter and enhancer histone marks, DNAase, changed motifs,
GRASP QTL hits, and bound proteins, suggesting GLS SNPs might produce
biological effects in lung cancer patients through these ways. The chi-square
test indicated that the genotype frequencies of these candidate SNPs met HWE
(p
| Gene | SNP ID | Functional annotation | Chr: position | Alleles (A/B) | MAF | HWE (p value) | OR (95% CI) | p | |
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||
| GLS | rs143584207 | missense_variant | 2:190,881,350 | C/A | 0.083 | 0.113 | 0.442 | 0.72 (0.55–0.92) | 0.010* |
| GLS | rs117985587 | 3’UTR | 2:190,913,888 | T/C | 0.057 | 0.054 | 0.129 | 1.05 (0.76–1.46) | 0.753 |
| GLS | rs74271715 | 3’UTR | 2:190,914,979 | G/T | 0.051 | 0.067 | 0.537 | 0.75 (0.54–1.03) | 0.075 |
| GLS | rs2355570 | 3’UTR | 2:190,916,443 | G/A | 0.497 | 0.465 | 0.218 | 1.14 (0.98–1.32) | 0.091 |
| GLS | rs6713444 | 3’UTR | 2:190,917,042 | A/G | 0.182 | 0.216 | 0.054 | 0.81 (0.67–0.97) | 0.024* |
| Notes: | |||||||||
In the case and control groups, the genotypes with the highest frequency distribution of rs143584207, rs117985587, rs74271715, rs2355570, and rs6713444 were AA, CC, TT, GA, and GG, respectively (Fig. 1). Rs143584207 could significantly reduce lung cancer susceptibility under the heterozygote (CA vs. AA: OR = 0.72, 95% CI 0.55–0.95, p = 0.020), additive (OR = 0.70, 95% CI 0.54–0.91, p = 0.008) and dominant (CC-CA vs. AA: OR = 0.70, 95% CI 0.54–0.92, p = 0.012) models. Rs6713444 could also significantly reduce lung cancer susceptibility under the heterozygote (AG vs. GG: OR = 0.78, 95% CI 0.62–0.98, p = 0.034), additive (OR = 0.79, 95% CI 0.65–0.96, p = 0.017) and dominant (AA-AG vs. GG: OR = 0.77, 95% CI 0.62–0.96, p = 0.021) models. There was no evidence of a marked correlation between the other three candidate SNPs (rs117985587, rs74271715, and rs2355570) and lung cancer susceptibility.
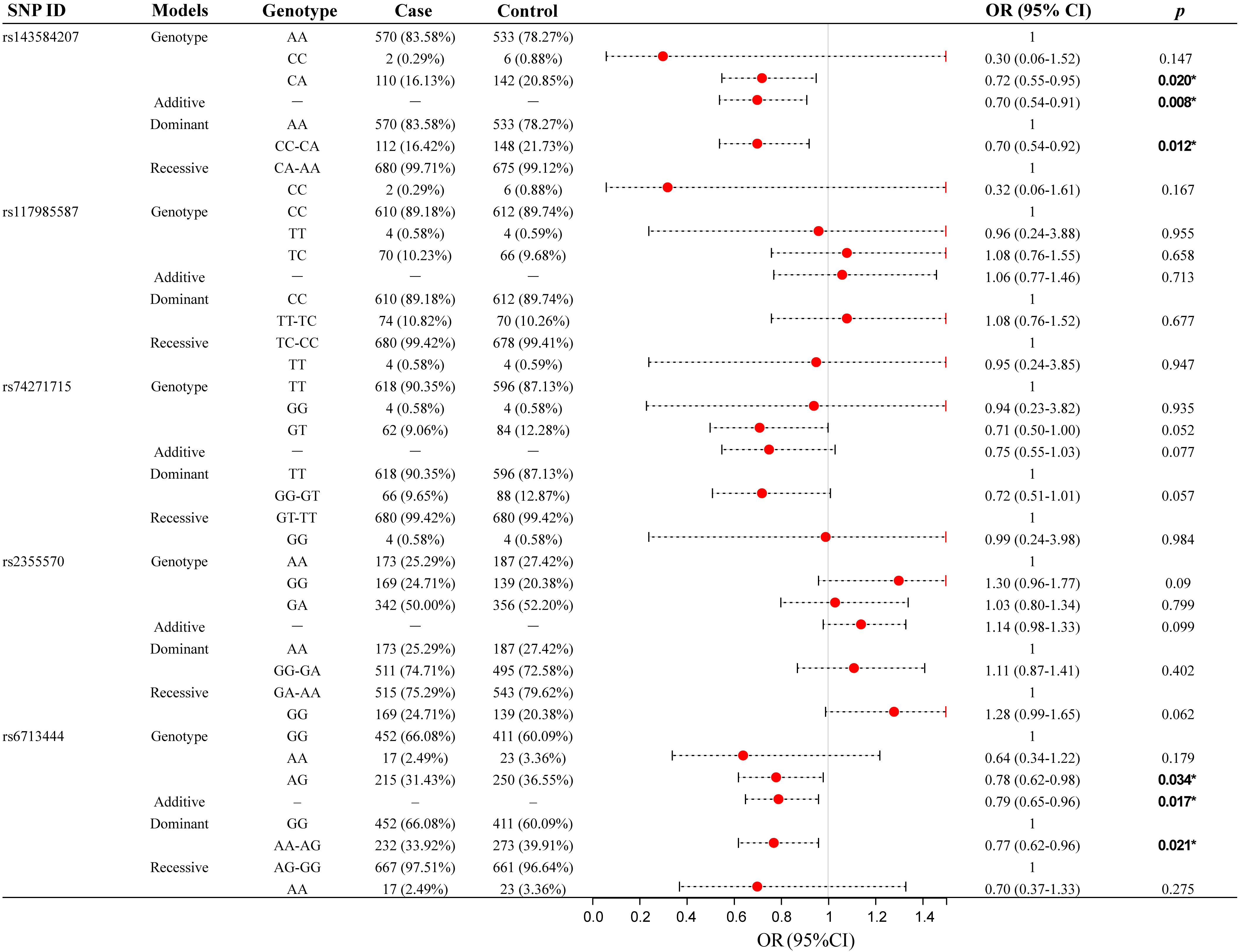 Fig. 1.
Fig. 1.Association between five candidate GLS SNPs and lung
cancer susceptibility (overall analysis). *p
Age-stratified analysis (Fig. 2, Supplementary Table 2) indicated that
there was no significant correlation between GLS SNPs and lung cancer
susceptibility in participants aged
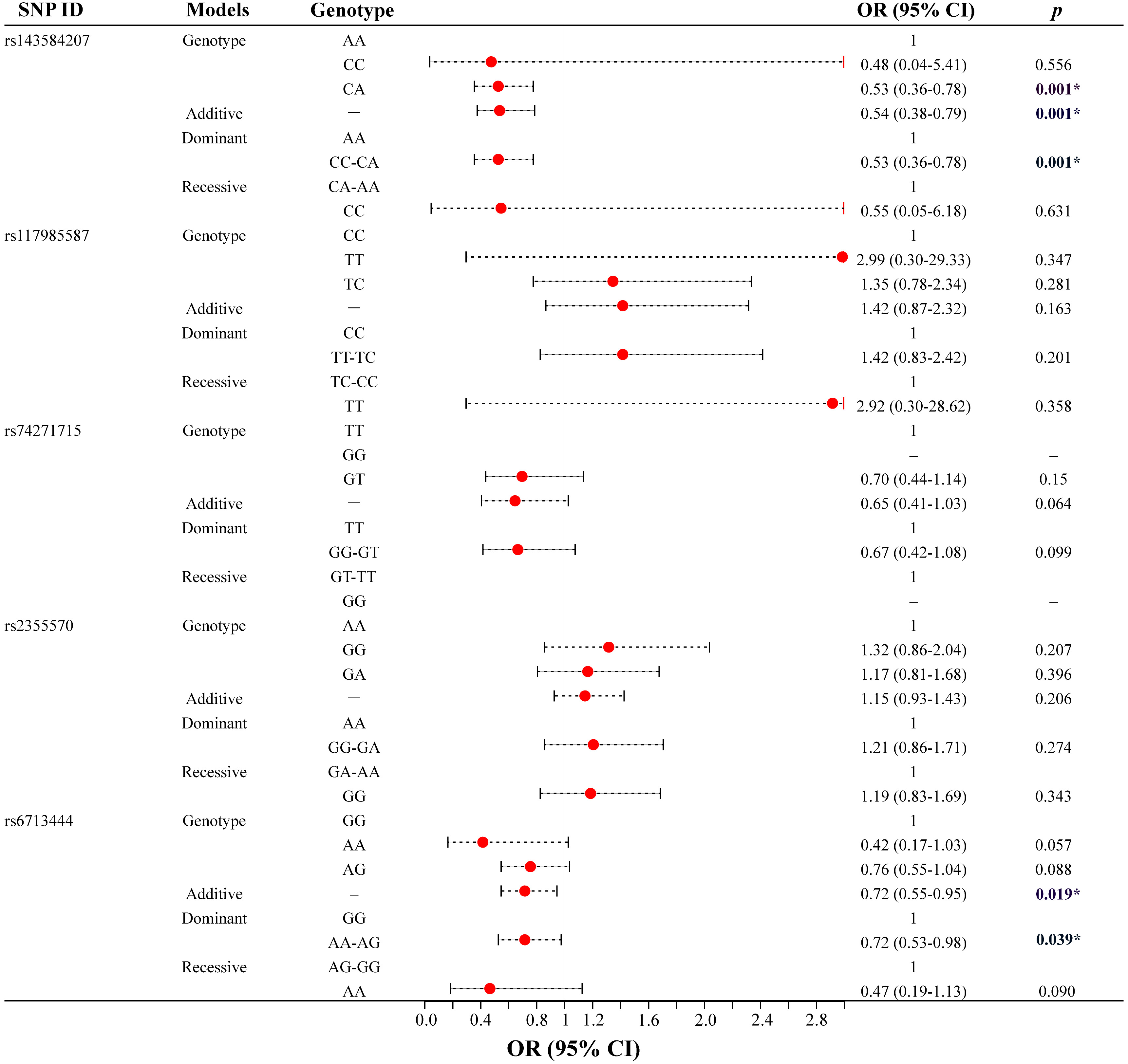 Fig. 2.
Fig. 2.Association between five candidate GLS SNPs and lung
cancer susceptibility (age-stratified analysis, age
Gender-stratified analysis (Supplementary Table 2) indicated that in female participants, rs143584207 could significantly reduce lung cancer susceptibility under the heterozygote (CA vs. AA: OR = 0.53, 95% CI 0.31–0.91, p = 0.021), additive (OR = 0.50, 95% CI 0.30–0.84, p = 0.008) and dominant (CC-CA vs. AA: OR = 0.51, 95% CI 0.30–0.86, p = 0.013) models, and rs6713444 could also significantly reduce lung cancer susceptibility under the heterozygote (AG vs. GG: OR = 0.58, 95% CI 0.38–0.89, p = 0.011), additive (OR = 0.67, 95% CI 0.47–0.95, p = 0.025) and dominant (AA-AG vs. GG: OR = 0.60, 95% CI 0.40–0.89, p = 0.012) models.
BMI-stratified analysis (Supplementary Table 3) revealed a marked
correlation of rs143584207 and rs6713444 with lung cancer susceptibility in
participants with BMI
Smoking-stratified analysis (Supplementary Table 3) showed that in non-smoking participants, only rs143584207 could significantly reduce lung cancer susceptibility under the additive (OR = 0.59, 95% CI 0.38–0.92, p = 0.020) and dominant (CC-CA vs. AA: OR = 0.61, 95% CI 0.39–0.96, p = 0.031) models. The other four genetic loci (rs117985587, rs74271715, rs2355570, and rs6713444) had no marked correlation with lung cancer susceptibility in non-smoking participants.
The clinical information about patients with different tumor types was analyzed, suggesting that (Fig. 3 and Supplementary Table 4) in patients with lung adenocarcinoma, those carrying the CA heterozygote of rs143584207 had a lower risk of lung adenocarcinoma than wild-type homozygous AA carriers (CA vs. AA: OR = 0.56, 95% CI 0.38–0.82, p = 0.003). And under the additive (OR = 0.53, 95% CI 0.36–0.76, p = 0.001) and dominant (CC-CA vs. AA: OR = 0.53, 95% CI 0.36–0.78, p = 0.001) models, rs143584207 could significantly reduce lung adenocarcinoma susceptibility. And rs6713444 could also significantly reduce lung adenocarcinoma susceptibility under the additive (OR = 0.66, 95% CI 0.51–0.86, p = 0.002) and dominant (AA-AG vs. GG: OR = 0.62, 95% CI 0.46–0.82, p = 0.001) models. However, in patients with squamous-cell lung cancer, we found that these five candidate SNPs of GLS had no significant correlation with squamous-cell lung cancer susceptibility.
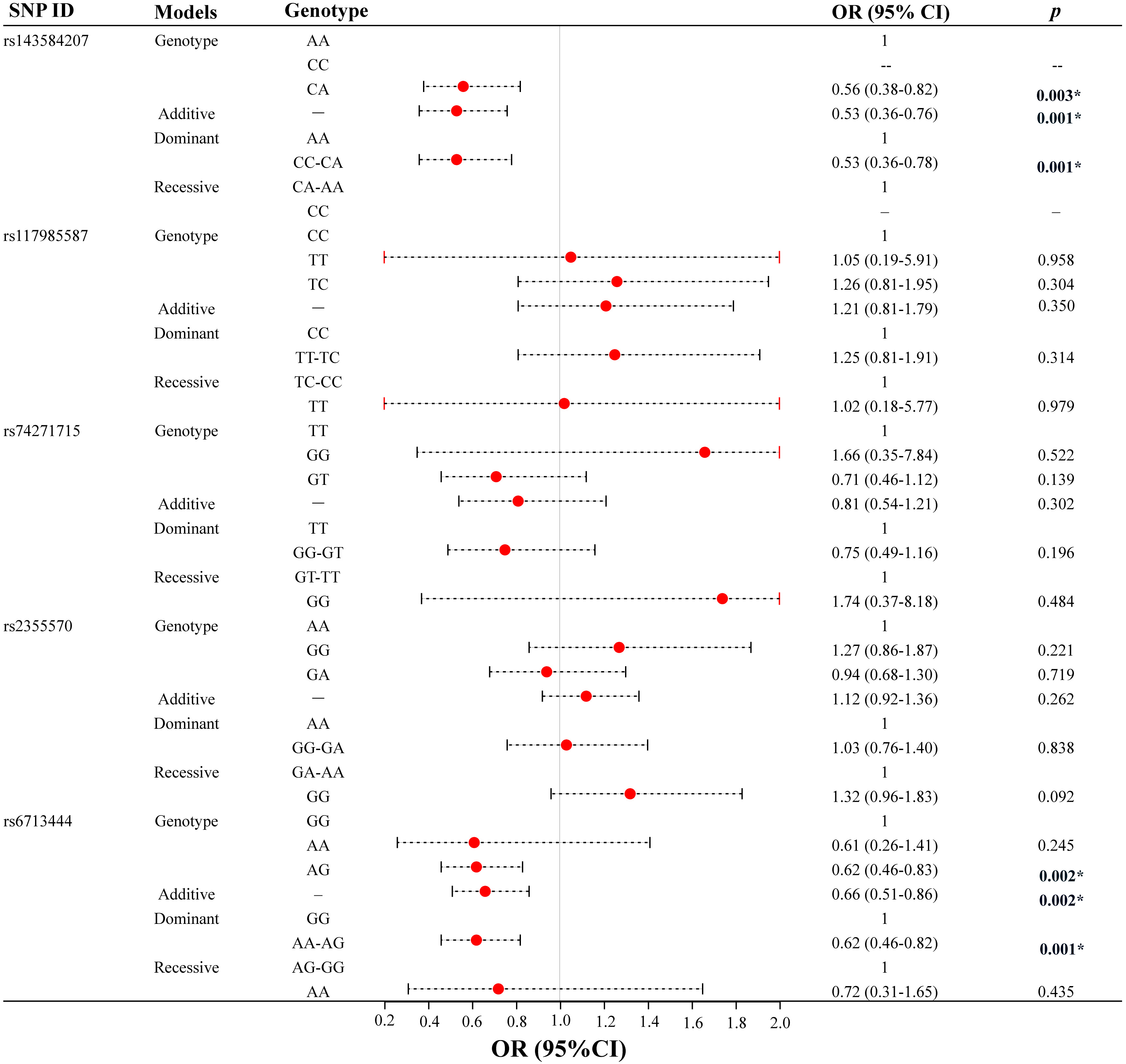 Fig. 3.
Fig. 3.Association between five candidate GLS SNPs and lung
cancer susceptibility (Lung adenocarcinoma, N = 314). *p
According to the four clinical stages of lung cancer, we took patients with stage I–II lung cancer as the control group. And the association between GLS SNPs and lung cancer susceptibility in patients with stage III-IV lung cancer was analyzed, as shown in Supplementary Table 5. Among patients with late-stage lung cancer, rs117985587 could significantly reduce lung cancer susceptibility under the additive (OR = 0.60, 95% CI 0.38– 0.95, p = 0.029) and dominant (TT-TC vs. CC: OR = 0.59, 95% CI 0.36–0.96, p = 0.035) models. However, rs6713444 was associated with a significantly increased susceptibility to lung cancer under the heterozygote (AG vs. GG: OR = 1.70, 95% CI 1.20–2.39, p = 0.003), additive (OR = 1.48, 95% CI 1.10–2.00, p = 0.011), and dominant (AA-AG vs. GG: OR = 1.64, 95% CI 1.18–2.28, p = 0.004) models.
When stratified by lymph node metastasis, we evaluated the association between GLS SNPs and lung cancer susceptibility, taking lung cancer patients without lymph node metastasis as the control group. No marked correlation between GLS SNPs and lung cancer susceptibility was observed in patients with lymph node metastasis (Supplementary Table 5).
The FPRP analysis of positive results of the overall and subgroup analyses was
further carried out, suggesting that (Supplementary Table 6) the
association between rs143584207 and lung cancer susceptibility should not be
concerned in non-smokers and participants with BMI
The interactions among five candidate SNPs of GLS were analyzed by the MDR method. As shown in Fig. 4, the blue lines indicate redundant interactions among the three candidate SNPs (rs6713444, rs74271715, and rs2355570). Additionally, MDR analysis (Table 3) showed that the two-locus model (the combination of rs143584207 and rs6713444) was considered as the best one (testing balanced accuracy = 0.5395, CVC = 9/10) to predict lung cancer susceptibility.
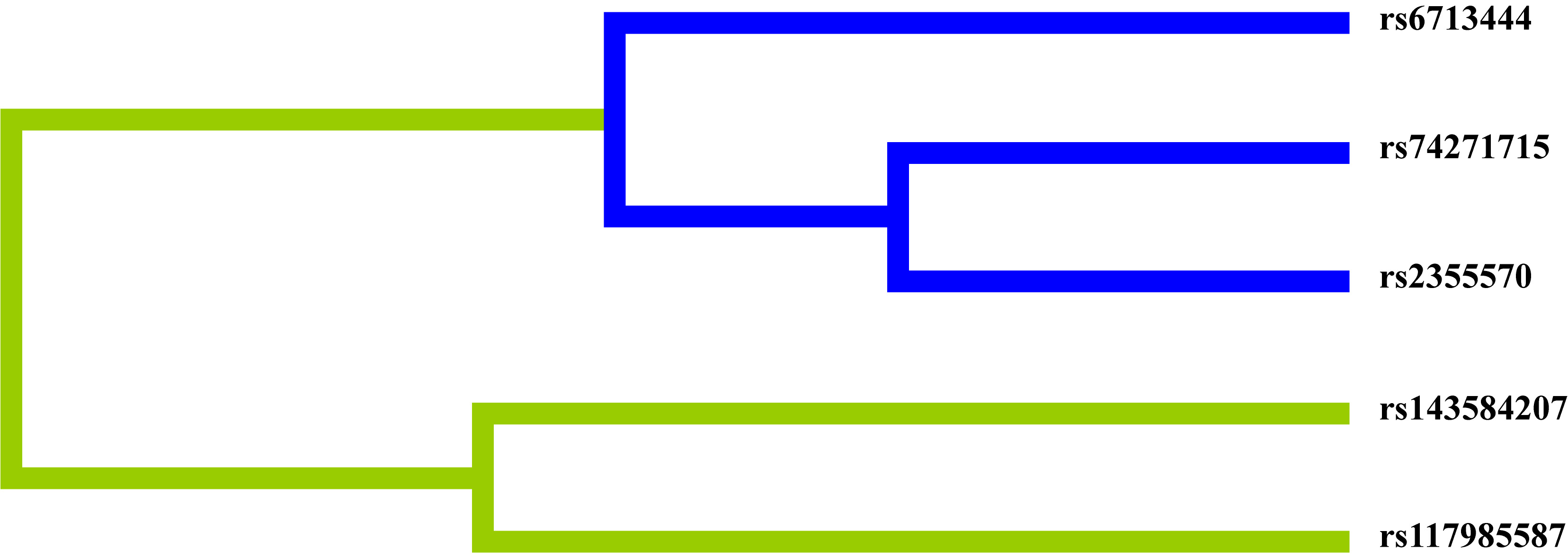 Fig. 4.
Fig. 4.Dendrogram of SNP-SNP interactions. Blue lines indicate redundancy between SNPs. Green lines indicate synergy between SNPs.
| Models | Training balanced accuracy | Testing balanced accuracy | OR (95% CI) | p | CVC |
|---|---|---|---|---|---|
| rs6713444 | 0.5307 | 0.5175 | 1.29 (1.04–1.61) | 0.0216 | 8/10 |
| rs143584207, rs6713444 | 0.5475 | 0.5395 | 1.46 (1.18–1.81) | 0.0004 | 9/10 |
| rs143584207, rs117985587, rs6713444 | 0.5530 | 0.5329 | 1.53 (1.23–1.89) | 8/10 | |
| rs143584207, rs117985587, rs74271715, rs6713444 | 0.5566 | 0.5336 | 1.56 (1.26–1.94) | 7/10 | |
| rs143584207, rs117985587, rs74271715, rs2355570, rs6713444 | 0.5608 | 0.5044 | 1.62 (1.31–2.01) | 10/10 | |
| Abbreviation: CVC, cross-validation consistency. | |||||
We analyzed the association between five candidate GLS SNPs and lung
cancer susceptibility among 1368 participants. The results indicated that
GLS rs143584207 and rs6713444 were significantly associated with lung
cancer susceptibility, while the other three SNPs (rs117985587, rs74271715, and
rs2355570) showed little correlation with lung cancer susceptibility. The overall
analysis revealed that GLS rs143584207 and rs6713444 might be protective
factors against lung cancer. The subgroup and clinical information analyses
further revealed that GLS rs143584207 and rs6713444 could significantly
reduce lung cancer susceptibility in different subgroups (age
Previous studies have found that SNPs in coding or non-coding areas may regulate gene function, gene expression level and enzyme activity by influencing the binding affinity of transcription factors and changing RNA splicing, thus significantly affecting tumor susceptibility [29]. Hu et al. [25] have found that the heterozygotes of rs1045411 can reduce the expression levels of the HMGB1 gene and thus decrease lung cancer susceptibility. Gemignani et al. [5] have detected that some gene polymorphisms encoding xenobiotic metabolizing enzymes (XMEs) are closely related to lung cancer susceptibility, and they may influence the levels of related metabolites and enzyme activities, thereby affecting lung cancer susceptibility. In this study, we preliminarily confirmed that GLS gene polymorphisms (rs143584207 and rs6713444) were markedly related to the reduction of lung cancer susceptibility. And we further speculated that rs143584207 and rs6713444 could reduce the expression of GLS, inhibit the hydrolysis of glutamine, affect the growth of tumor cells, and ultimately decrease lung cancer susceptibility. But the specific mechanism of GLS SNPs in lung cancer development needs to be further demonstrated by follow-up study.
In addition, more and more studies have shown that smokers and non-smokers are both susceptible to lung cancer, nevertheless, their various clinicopathologic features may suggest that the etiologies of lung cancer in smokers and non-smokers are different [30]. Noticeably, some gene polymorphisms may have a marked impact on lung cancer susceptibility [31, 32]. Li et al. [9] have found that genetic variants on chromosome 13q31.3 can reduce the expression of GPC5 and are related to an increased lung cancer susceptibility in non-smokers. Jou et al. [33] have discovered that one SNP (8227G) located in the intron of EGFR has a major impact on increased lung cancer susceptibility, especially in non-smoking female patients with lung adenocarcinoma. Hence, we thought that SNPs in GPC5 and EGFR might be risk factors for lung cancer in non-smokers. In contrast, Zhang et al. [32] have noticed that AGBL1 rs4513061 can reduce lung cancer susceptibility in non-smoking females. Our study also found that GLS rs143584207 could significantly reduce lung cancer susceptibility in non-smoking females. Therefore, we considered that AGBL1 rs4513061 and GLS rs143584207 might be protective factors against lung cancer in non-smoking females. These findings can give an important direction for the timely treatment of lung cancer among non-smoking females.
What’s more, our research also showed that the association between GLS
SNPs and lung cancer susceptibility was affected by age, sex, and pathological
types. GLS rs143584207 and rs6713444 could significantly reduce the
susceptibility of lung cancer in participants aged
It is undeniable that there are some shortcomings in this research. The sample size was relatively small, and all subjects were recruited from the same hospital, which cannot fully represent the Chinese Han population. We will further expand the sample size in follow-up studies so as to obtain more accurate and convincing results.
Our study found that GLS rs143584207 and rs6713444 could significantly reduce lung cancer susceptibility in the Chinese Han population, which will give a new direction for the timely treatment of lung cancer in high-risk populations.
SNPs, single nucleotide polymorphisms; GLS, glutaminase; ORs, odds ratios; 95% CIs, 95% confidence intervals; FPRP, false-positive report probability; MDR, multi-factor dimensionality reduction; BMI, body mass index; NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; VEGF, vascular endothelial growth factor; IL-32, interleukin-32; PARK2, parkin RBR E3 ubiquitin protein ligase; KGA or GLS1, kidney-type glutaminase; LGA or GLS2, liver-type glutaminase; TNM, tumor, node, metastasis; HWE, Hardy-Weinberg equilibrium; MAF, minor allele frequency; XMEs, xenobiotic metabolizing enzymes.
The datasets generated and/or analyzed during this study are available from the corresponding author on reasonable request.
YW and MingyC designed the study; YW, FY, and JX performed the research; CL, ZZ, and PW analyzed the data; YW, TJ, and MingwC wrote, reviewed, and edited the manuscript. All authors have participated sufficiently in the work and agreed to be responsible for all aspects of this work. All authors have contributed to editorial changes in the manuscript and have read and approved the final manuscript.
This study was conducted under the standards approved by the Biomedical Ethics Committee of Xizang Minzu University (No. 20200-11), and conformed to the ethical principles of the World Medical Association Declaration of Helsinki for medical research involving humans. All participants signed informed consent forms before participating in this study.
We thank all authors and participants for their support and contributions.
This research received no external funding.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
