1 Jiangsu Key Laboratory of Oral Diseases, Nanjing Medical University & Department of Endodontic, Affiliated Hospital of Stomatology, Nanjing Medical University, 211166 Nanjing, Jiangsu, China
2 Institute of Stomatology, Nanjing Medical University, 211166 Nanjing, Jiangsu, China
3 Department of Endodontics, Stomatological Hospital, School of Stomatology, Southern Medical University, 510280 Guangzhou, Guangdong, China
4 Department of Oral and Maxillofacial Surgery, Stomatological Hospital, School of Stomatology, Southern Medical University, 510280 Guangzhou, Guangdong, China
Abstract
Background: Human dental stem cells (DSCs) are excellent sources of cells for treating dental and craniofacial diseases. However, the mechanisms regulating DSC osteogenic differentiation are still unclear. In this study, we aimed to determine the role of Krüppel-like factor 9 (KLF9) in regulating the biological functions of DSCs and explore the underlying molecular mechanisms. Methods: Bioinformatic analyses, quantitative real-time polymerase chain reaction (qRT‒PCR) and Western blotting were performed to determine the KLF9 level during osteogenic differentiation of DSCs. The effects of KLF9 depletion or overexpression on DSC osteogenic differentiation were then evaluated. The osteogenic potential and associated mineralized nodule-forming activities of DSCs were monitored via Alizarin red S staining and quantitative analyses of osteogenic markers. The regulatory effect of KLF9 on the Notch1 signaling pathway was analyzed by luciferase reporter assays. Results: KLF9 mRNA expression was consistently increased during mesenchymal stem cell osteogenic differentiation in multiple public datasets, and our qRT‒PCR and Western blotting data further validated this finding. In addition, KLF9 depletion promoted proliferation and suppressed osteogenic differentiation of DSCs, while enforced expression of KLF9 promoted the DSC osteogenic potential. Mechanistically, KLF9 negatively regulated the Notch1-mediated signaling pathway by directly binding to the Notch1 promoter. More importantly, Notch1 inhibition/overexpression partially rescued the suppressive/enhancing effects of KLF9 depletion/overexpression on the osteogenic differentiation of DSCs, indicating that Notch1 is a functional downstream target of KLF9. Conclusions: In summary, our results strongly demonstrate that KLF9 is a crucial transcription factor that controls the osteogenic differentiation of DSCs by negatively regulating the Notch1 signaling pathway.
Keywords
- dental stem cells
- KLF9
- osteogenic differentiation
- Notch1
- signaling pathway
Bone defects in the craniomaxillofacial region may occur due to trauma, infection, surgical excision, periodontal diseases and congenital deformities. Restoration of these defects is extremely difficult due to the complex anatomical structures [1, 2]. Stem cells are a class of primitive cells that show self-renewal and multidirectional differentiation. Stem cell-based tissue regeneration is an attractive approach for regenerating injured or pathologically damaged tissues [3, 4]. To date, there are at least six various types of dental stem cells (DSCs) [5]. It should be pointed out that different types of dental tissue-derived mesenchymal stem cells might possess the same phenotype. However, significant differences in the differentiation ability and expression of molecular markers were found among distinct DSCs [6]. Various strategies have been adopted to enhance the osteogenic differentiation of DSCs [7, 8]. For instance, Chan et al. [9] reported that three-dimensional culture markedly enhanced the stemness and differentiation potential of human dental pulp stem cells (hDPSCs). Controlling the commitment of DSCs into the osteogenic lineage is crucial for the success of hard tissue regeneration [10]. Thus, elucidating the molecular mechanisms underlying DSC osteogenic differentiation is urgently needed.
Krüppel-like factor 9 (KLF9) was initially isolated from a liver cDNA
library [11]. It has been demonstrated that KLF9 plays a critical role in various
physiological processes such as cell proliferation, differentiation and
development [12]. KLF9 was shown to promote neuronal cell differentiation, B-cell
differentiation, and endometrial cell proliferation [13, 14, 15]. In addition, KLF9
was reported to be a key transcription factor regulating the morphological
differentiation of oligodendrocytes, and it was induced in differentiated
oligodendrocytes by triiodothyronine [16]. KLF9 acted synergistically with
C/EBP
Our internal RNA-seq data indicated that KLF9 expression was markedly upregulated during osteogenic differentiation of human periodontal ligament stem cells (hPDLSCs) and human dental pulp stem cells (hDPSCs), indicating that it might be a crucial regulator for modulating DSC osteogenic differentiation. In this study, we demonstrated that KLF9 was consistently upregulated in DSCs undergoing osteogenic induction. In addition, KLF9 depletion promoted the proliferation and suppressed the osteogenic differentiation of DSCs. Ectopic expression of KLF9 enhanced the osteogenic potential of DSCs. Mechanistically, KLF9 negatively regulated the Notch1 signaling pathway by directly binding to the promoter of Notch1. Enforced expression of Notch1 partially decreased the promoting effects of KLF9 overexpression on DSC osteogenic differentiation, and vice versa.
This study was approved by the Ethics Committee of Nanjing Medical University
and conformed to the ethical principles of the Declaration of Helsinki. Patients
enrolled in the study provided written informed consent for the use of their
tissues. The hDPSCs and hPDLSCs were isolated and cultured as we previously
described [8, 20]. The DSCs were maintained in complete growth media at 37
°C with 5% CO
Short hairpin RNAs (shRNAs) targeting the KLF9 gene were inserted into the LV3-pGLV-h1-GFP-puro vector (GenePharma, Shanghai, China) to produce shKLF9. For generation of KLF9 or Notch1 overexpression plasmids, the full-length human KLF9 gene or Notch1 gene was cloned into a pGCL-GFP vector (GeneChem, Shanghai, China). The packaging plasmids and recombinant lentiviral vectors were cotransfected into HEK-293T cells to generate lentiviruses. The lentiviruses were collected and concentrated after 72 h of incubation. The DSCs were then transduced with the filtered lentiviruses at a multiplicity of infection of 50. The lentiviral particles targeting Notch1 gene were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
The seeding intensity of MSCs was 3
The cells with the indicated treatments were incubated with 10
The total RNA was extracted with a Quick-RNA™ kit (Zymo Research
Corp, Irvine, CA, USA). The first-strand cDNA was synthesized with
SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen).
Relative mRNA quantification was determined with SYBR Green I MasterMix (Roche,
Basel, Switzerland) on a CFX Connect Real-Time PCR Detection System (Bio-Rad,
Hercules, CA, USA). Each sample was measured at least three times. The relative
mRNA levels were determined with the 2
Equal amount of proteins were loaded onto 4–20% SDS‒PAGE gels (Beyotime, Shanghai, China) and subjected to electrophoresis. PVDF membranes (Bio-Rad) were activated in methanol, and electrophoretic transfer was performed with the Trans-Blot Turbo Transfer system (Bio-Rad). The membranes were then immersed in fast-blocking buffer (EpiZyme, Shanghai, China) for 10 min, and probed with the indicated primary antibodies overnight in cold room. After being washed with TBST buffer, the membranes were incubated with secondary antibodies for 1 h at room temperature. The primary antibodies were listed as follows: anti-KLF9 (Abcam, Cambridge, UK), anti-GAPDH (Proteintech, Chicago, IL, USA), anti-ALP (Abcam), anti-Osx (Thermo Fisher Scientific, Waltham, MA, USA), anti-Dlx2 (Thermo Fisher Scientific), anti-BMP2 (Proteintech), anti-Runx2 (Proteintech), anti-COL1A1 (Proteintech), anti-Notch1 (Abcam), anti-HES1 (Thermo Fisher Scientific), and anti-HEY1 (Proteintech).
The datasets evaluating dynamic mRNA expression during MSC osteogenic differentiation were obtained from the Gene Expression Omnibus database. The accession numbers were GSE18072, GSE49007, GSE80614, GSE145813 and GSE112694.
The DSCs with the indicated treatments were fixed in 4% paraformaldehyde and stained with Alizarin red S solution (Cyagen, Santa Clara, CA, USA) for 10 min at 25 °C.
The indicated cells were transfected with Notch1 promoter vectors or mutant plasmids with the Lipofectamine 3000 transfection reagent (Invitrogen). After 24 h of incubation, a Dual-Luciferase Assay System (Promega, Madison, WI, USA) was used to measure the relative luciferase activities according to the manufacturer’s protocol.
The GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA) was used to
perform the statistical analyses. Data were presented as the mean
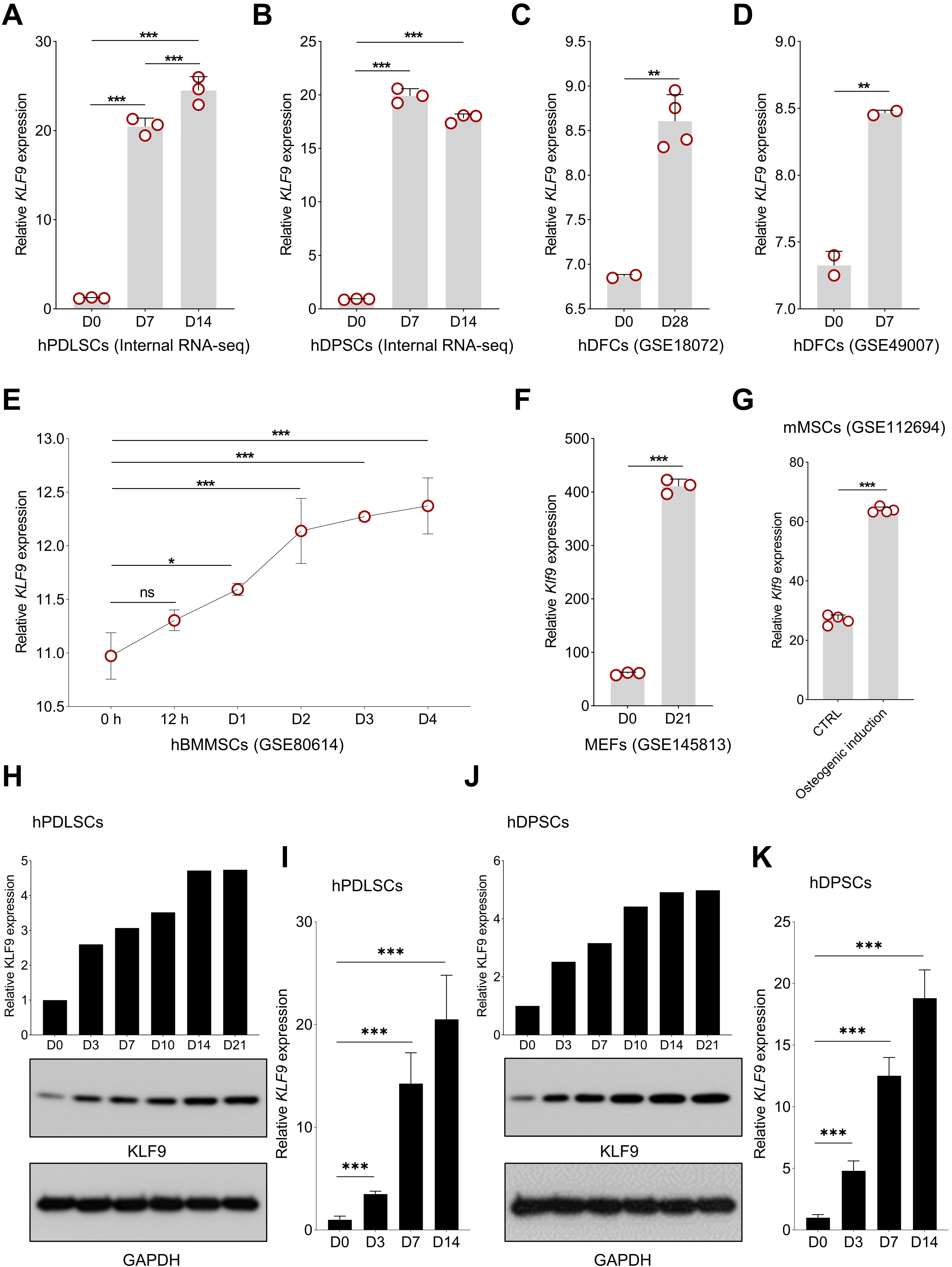
Our internal RNA-seq data revealed that KLF9 expression was consistently increased during osteogenic differentiation of hPDLSCs and hDPSCs (Fig. 1A,B). Then, we further evaluated the expression profiles of KLF9 during osteogenic differentiation with public assessable datasets. As shown in Fig. 1C,D, KLF9 was significantly upregulated in the human dental follicle cells (DFCs) with osteogenic induction. Interestingly, the expression of KLF9 was also gradually upregulated at the early stage of human bone marrow MSC osteogenic differentiation (Fig. 1E). Klf9 expression was also markedly increased during osteogenic differentiation of mouse stem cells (Fig. 1F,G). Our Western blotting and qRT‒PCR results further confirmed that KLF9 protein and mRNA were significantly increased during osteogenic differentiation of hPDLSCs (Fig. 1H,I). Similar findings were observed in hDPSCs with osteogenic induction (Fig. 1J,K).
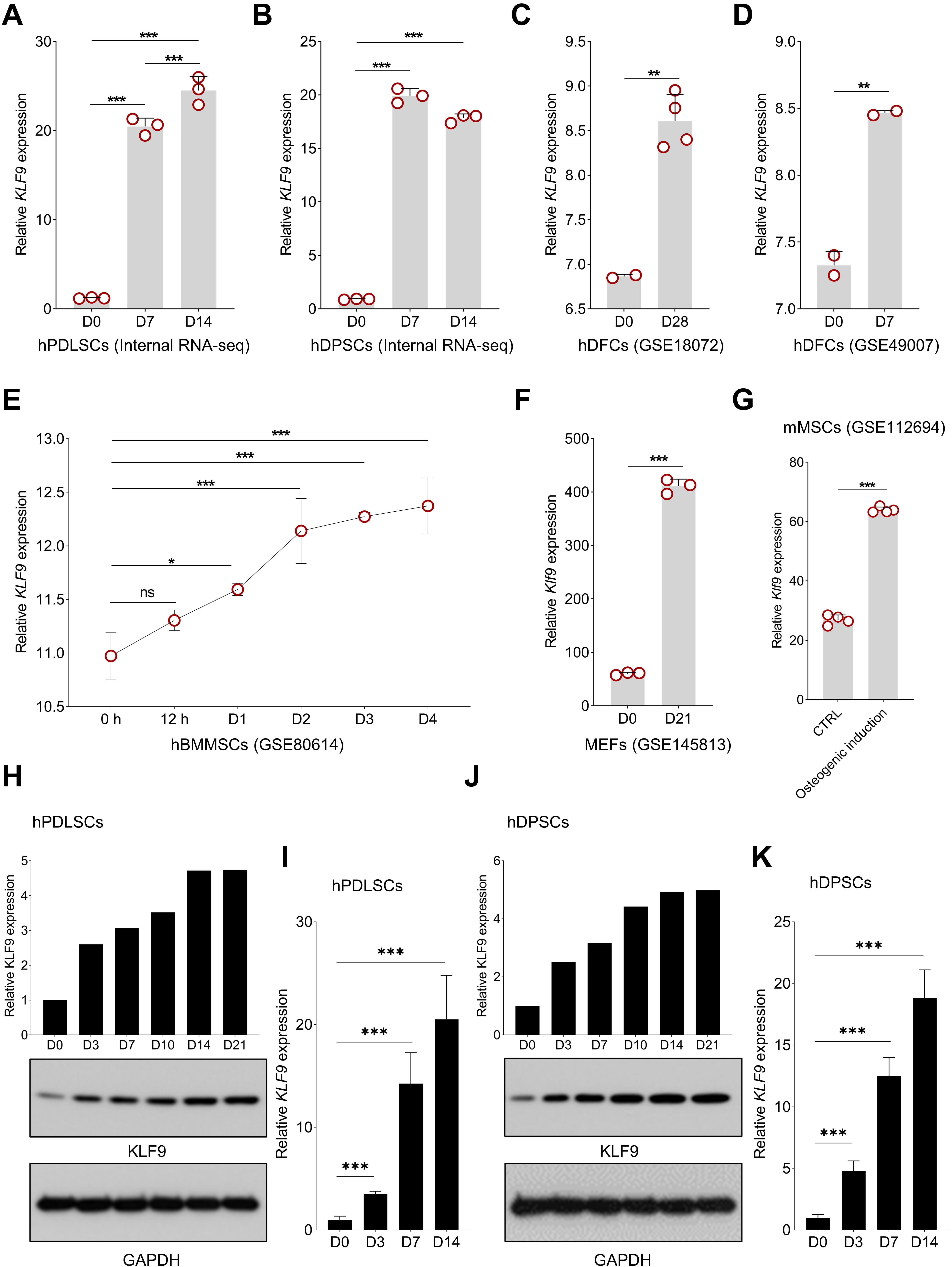 Fig. 1.
Fig. 1.KLF9 expression is increased during MSC osteogenic
differentiation. (A,B) The expression pattern of KLF9 during the
osteogenic differentiation of hPDLSCs and hDPSCs based on our internal RNA-seq
data. (C,D) The alteration of KLF9 level during osteogenic
differentiation of hDFCs in GSE18072 and GSE49007. (E) The dynamic expression
profile of KLF9 at the early stage of hBMMSC osteogenic differentiation
in GSE80614. (F,G) Changes in Klf9 expression in mouse stem cells with
osteogenic induction in GSE145813 and GSE112694. (H) The dynamic change of KLF9
protein in hPDLSCs with osteogenic induction at the indicated time points. (I)
The expression of KLF9 mRNA during osteogenic differentiation of hPDLSCs. (J) The
dynamic change of KLF9 protein in hDPSCs with osteogenic induction at the
indicated time points. (K) qRT‒PCR analysis of KLF9 mRNA expression during
osteogenic differentiation of hDPSCs. *p
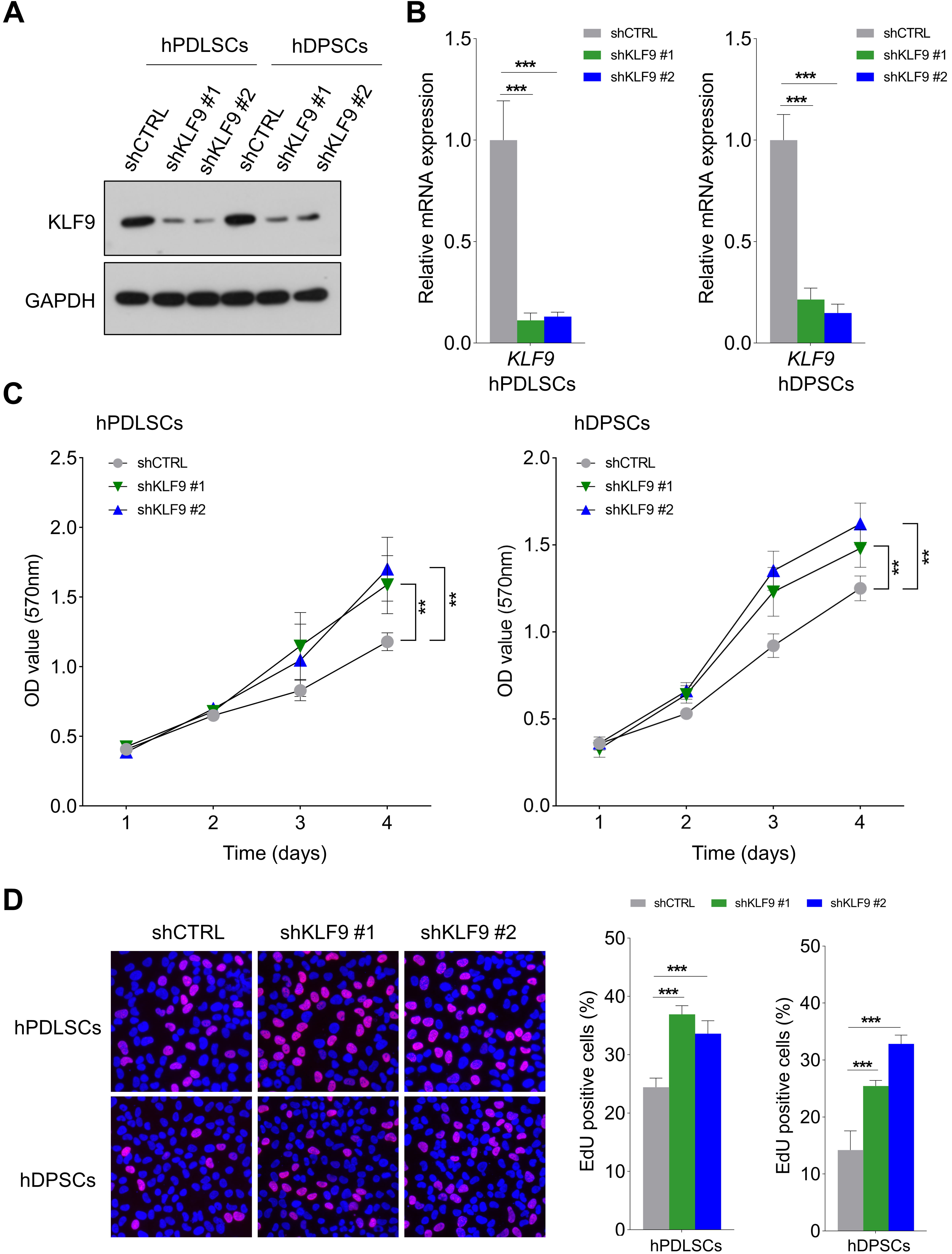
As shown in Fig. 2A,B, the shRNAs targeting KLF9 could effectively reduce the expression of KLF9 at both the protein and mRNA levels in hPDLSCs and hDPSCs. Then, the effects of KLF9 depletion on the proliferation of DSCs were evaluated. As revealed by the MTT assay, the OD values were noticeably higher in the shKLF9-treated cells (Fig. 2C). Similarly, the percentage of cells positive for EdU was markedly higher in the KLF9 knockdown groups (Fig. 2D).
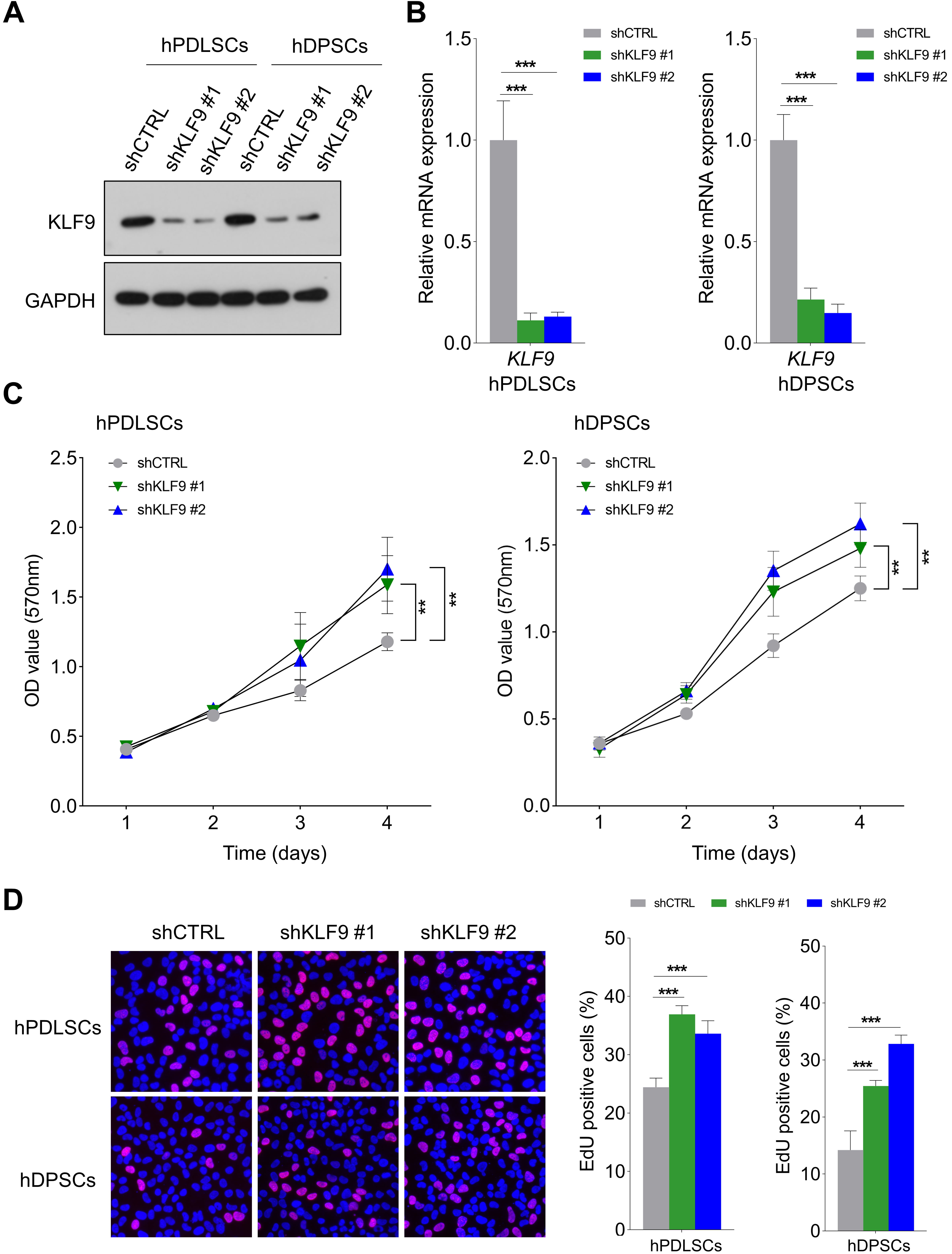 Fig. 2.
Fig. 2.KLF9 depletion promotes the proliferation of DSCs. (A,B) The
shRNAs targeting KLF9 efficiently reduced the expression of KLF9 protein and mRNA
in DSCs. (C) MTT assays of the optical density (OD) values of KLF9-depleted cells and control
cells. (D) The percentage of cells positive for EdU in the KLF9 depletion groups
and the control group. **p
The effects of KLF9 depletion or overexpression on DSC osteogenic differentiation were further determined. The mRNA levels of osteogenic markers were significantly lower in the KLF9-depleted cells than in the control cells (Fig. 3A,B). Similarly, KLF9 depletion markedly reduced the expression of these osteogenic markers at the protein level (Fig. 3C,D). Alizarin red S staining demonstrated that the mineralized nodule formation was significantly reduced in the DSCs with KLF9 depletion compared to the DSCs with normal KLF9 expression (Fig. 3E,F).
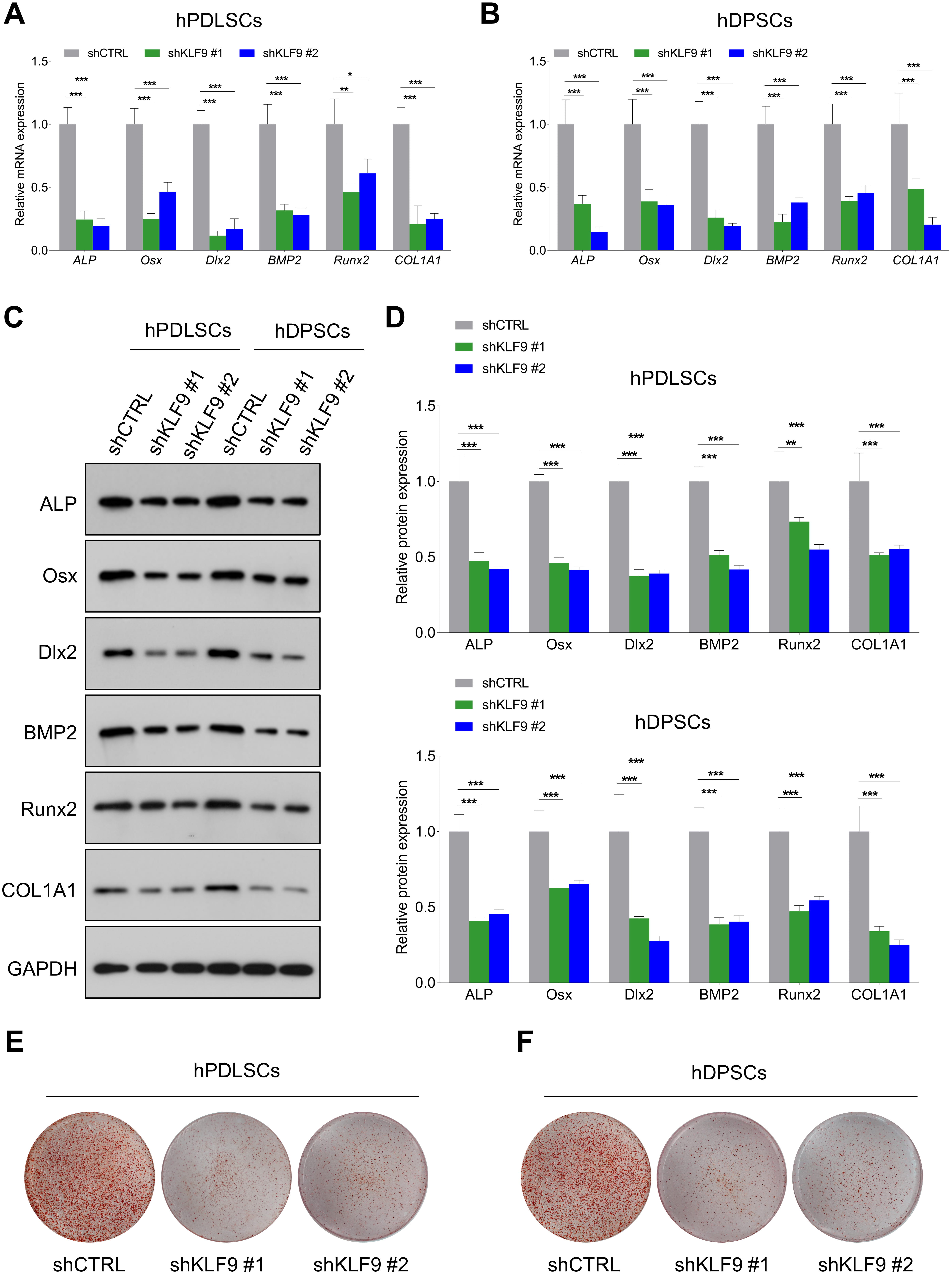 Fig. 3.
Fig. 3.KLF9 depletion suppresses DSC osteogenic differentiation. (A,B)
After osteogenic differentiation for 14 days, qRT-PCR was performed to assess the
mRNA levels of osteogenic markers in KLF9 depleted cells and the control cells.
(C,D) After osteogenic differentiation for 14 days, western blotting was
conducted to determine the expression of osteogenic markers in KLF9 depleted
cells and the control cells. (E,F) After osteogenic differentiation for 21 days,
the mineralized nodule forming capacities of KLF9-depleted cells and the control
cells were assessed by alizarin red S staining. *p
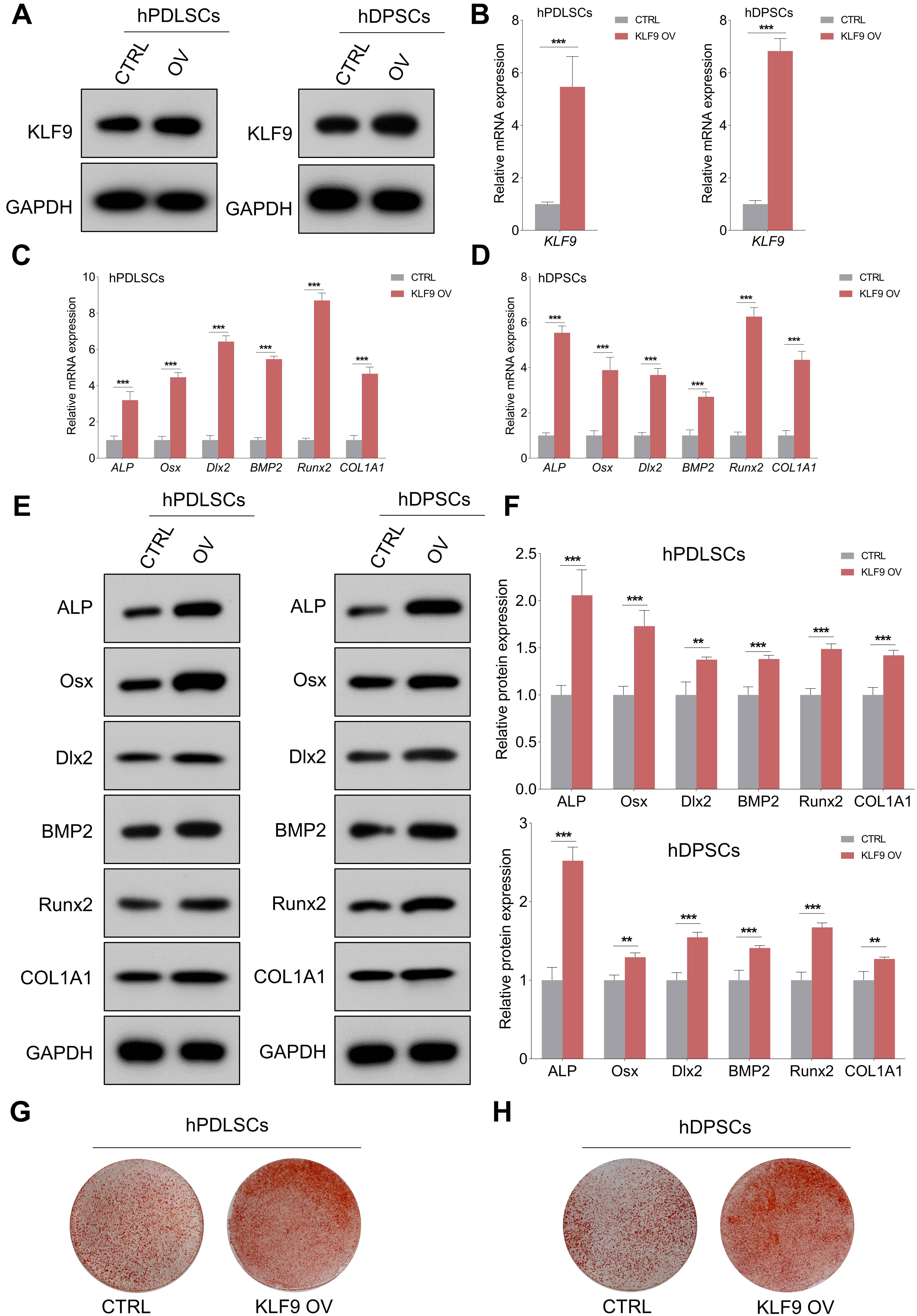
The expression of KLF9 protein and mRNA was efficiently upregulated in DSCs transduced with KLF9 overexpression lentiviruses (Fig. 4A,B). As expected, enforced expression of KLF9 markedly increased the expression of ALP, Osx, Dlx2, BMP2, Runx2 and COL1A1 compared to that of the control cells (Fig. 4C–F). Alizarin red S staining revealed that the KLF9-overexpressing cells had significantly higher mineralized nodule formation than the control cells (Fig. 4G,H).
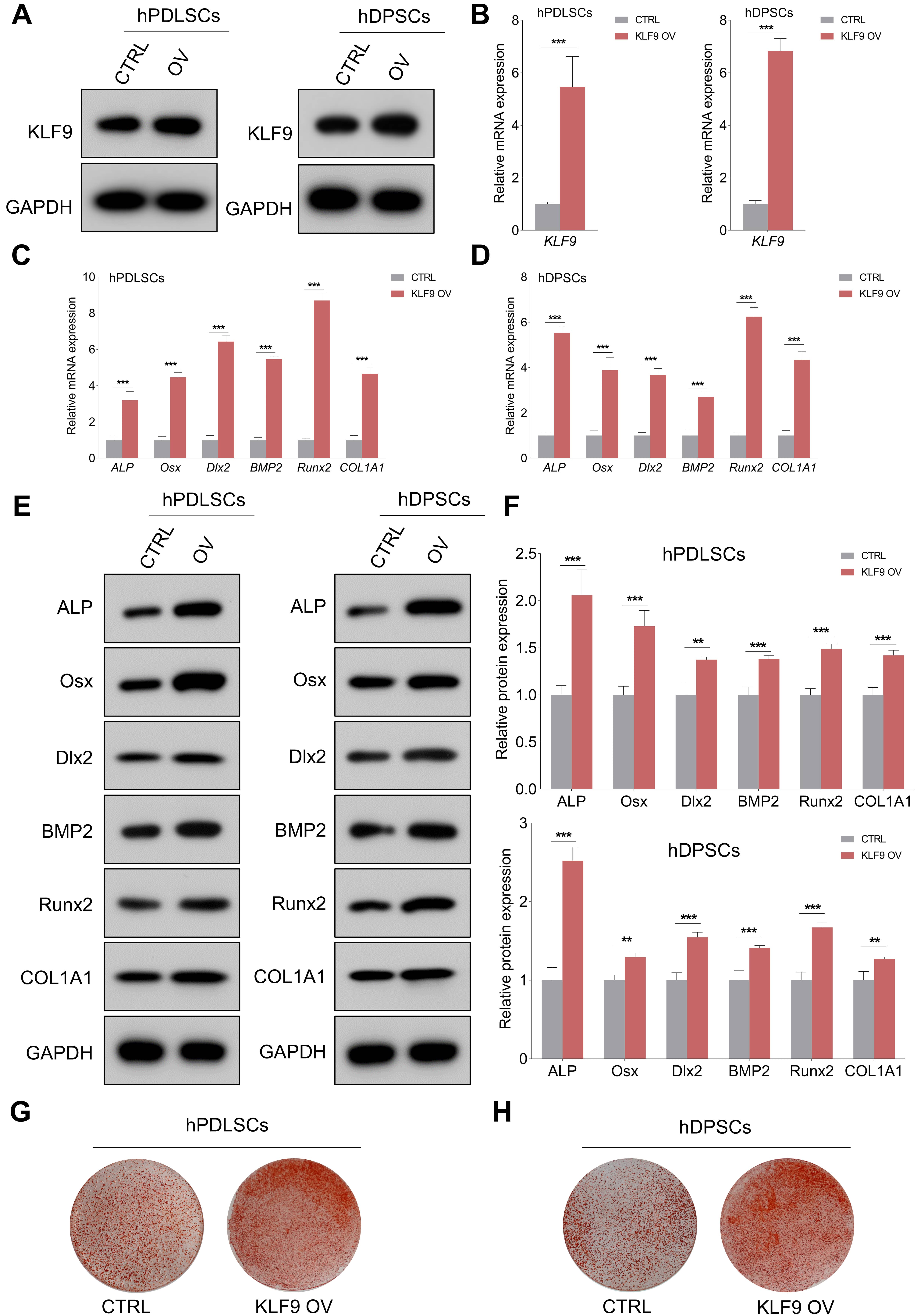 Fig. 4.
Fig. 4.KLF9 overexpression enhances osteogenic differentiation of
DSCs. (A,B) KLF9-overexpressing lentiviruses significantly elevated the
expression of KLF9 protein and mRNA in DSCs. (C,D) After osteogenic
differentiation for 14 days, qRT-PCR was performed to evaluate the expression of
osteogenic markers in KLF9 overexpressing cells and the control cells. (E,F)
After osteogenic differentiation for 14 days, western blotting was performed to
analyze the expression of osteogenic markers in KLF9 overexpressing cells and the
control cells. (G,H) After osteogenic differentiation for 21 days, the
mineralized nodule forming capacity of DSCs with indicated treatments was
assessed with alizarin red S staining. **p
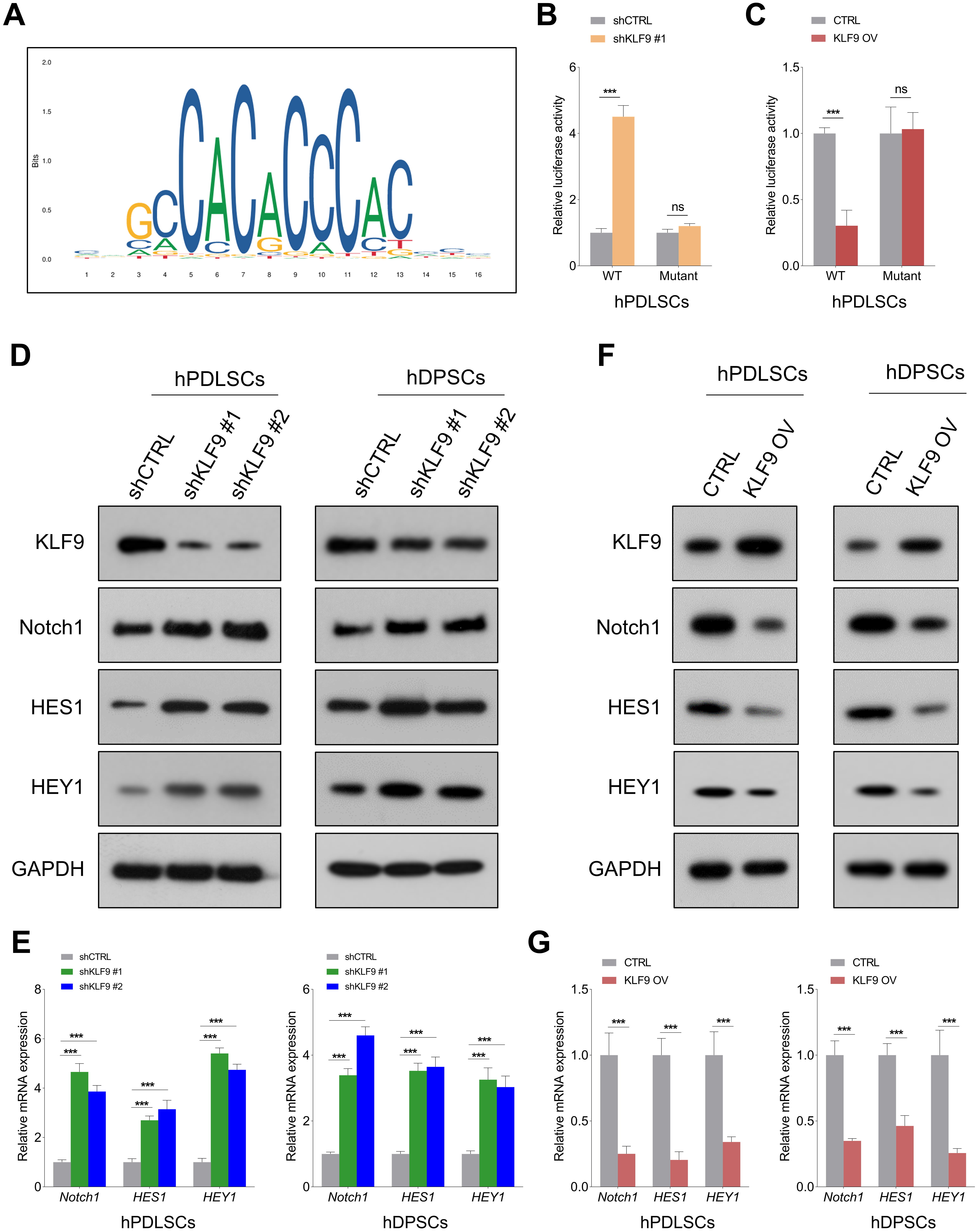
Notch1 signaling is crucial for modulating the osteogenic differentiation of DSCs [21], and KLF9 is an important upstream regulator of Notch1 signaling [22]. To determine the potential underlying mechanism of KLF9 in mediating Notch1 transcription, we predicted the binding sequences of KLF9 using the JASPAR database (Fig. 5A). Then, wild-type and mutant reporter vectors containing KLF9 binding sites (ACCCCCACCCACC) were constructed. The luciferase reporter assay revealed that KLF9 depletion markedly enhanced Notch1-luciferase activity, and KLF9 overexpression led to the opposite findings. Interestingly, KLF9 upregulation/downregulation had little effect on the luciferase activities of the mutant Notch1 promoter (Fig. 5B,C, Supplementary Fig. 1A,B). In addition, the Western blotting and qRT‒PCR results showed that KLF9 depletion significantly increased the expression of Notch1 and its downstream targets (HES1, HEY1) in DSCs, and ectopic expression of KLF9 suppressed Notch1-mediated signaling (Fig. 5D–G, Supplementary Fig. 2A–D).
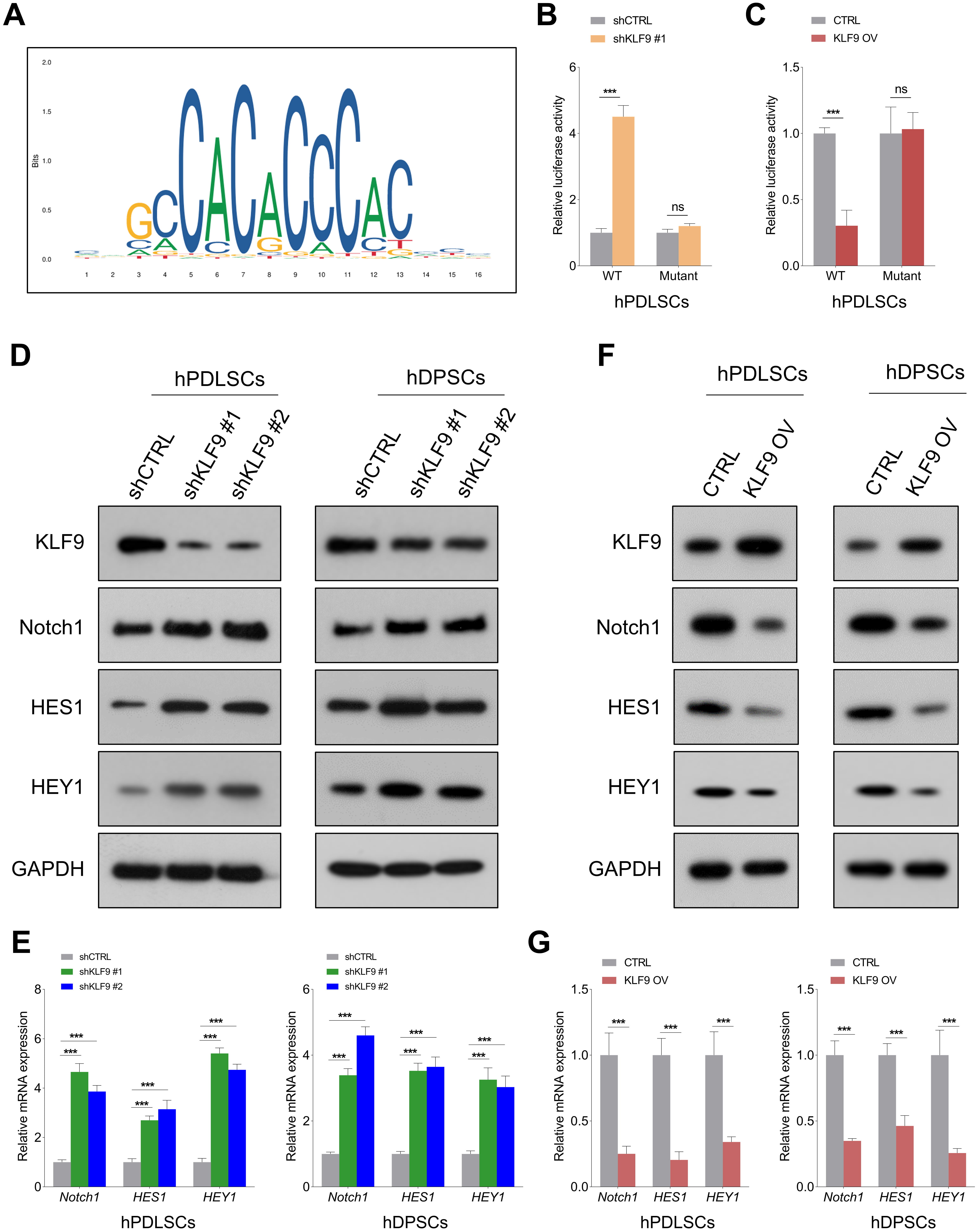 Fig. 5.
Fig. 5.KLF9 negatively regulates the Notch1-mediated signaling pathway
in DSCs. (A) Prediction of the KLF9 sequence logo with the JASPAR database.
(B,C) The relative luciferase activities in hPDLSCs with the indicated
treatments. (D,E) The expression of Notch1 and its downstream targets (HES1 and
HEY1) at both the protein and mRNA levels in KLF9-depleted cells and control
cells. (F,G) The expression of Notch1, HES1 and HEY1 in KLF9-overexpressing cells
and control cells. ns, not significant, ***p
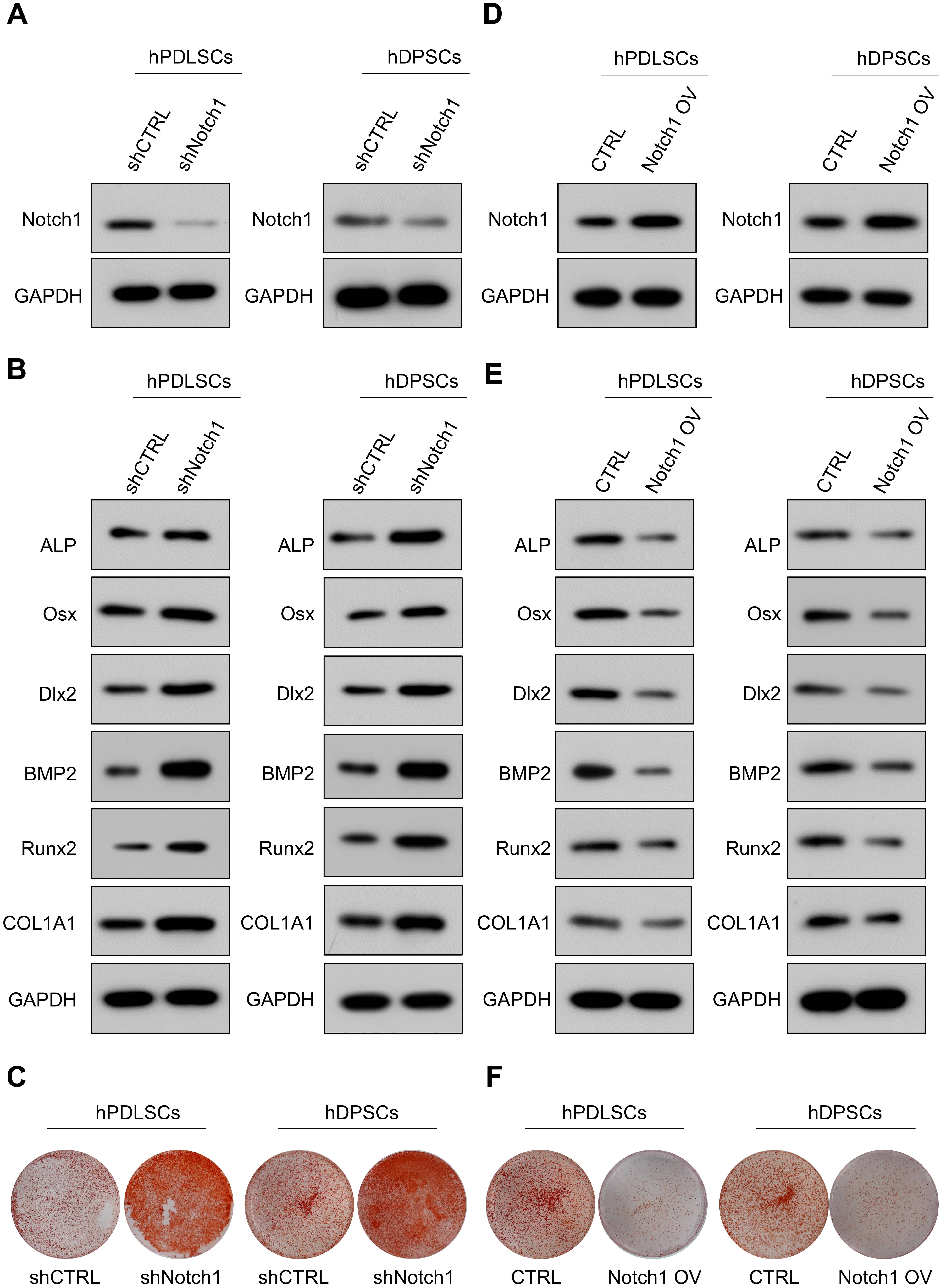
We then explored the effects of Notch1 on DSC osteogenic differentiation. As expected, Notch1 depletion significantly promoted the expression of osteogenic markers (Fig. 6A,B, Supplementary Fig. 3A–C). In addition, the mineralized nodule formation was significantly promoted in the Notch1-depleted cells (Fig. 6C). Similarly, Notch1 overexpression markedly reduced the levels of osteogenic markers in DSCs (Fig. 6D,E, Supplementary Fig. 3D–F). Enforced expression of Notch1 significantly reduced the mineralized nodule formation in DSCs (Fig. 6F). These findings strongly indicated that Notch1 acted as a negative regulator for modulating DSC osteogenic differentiation.
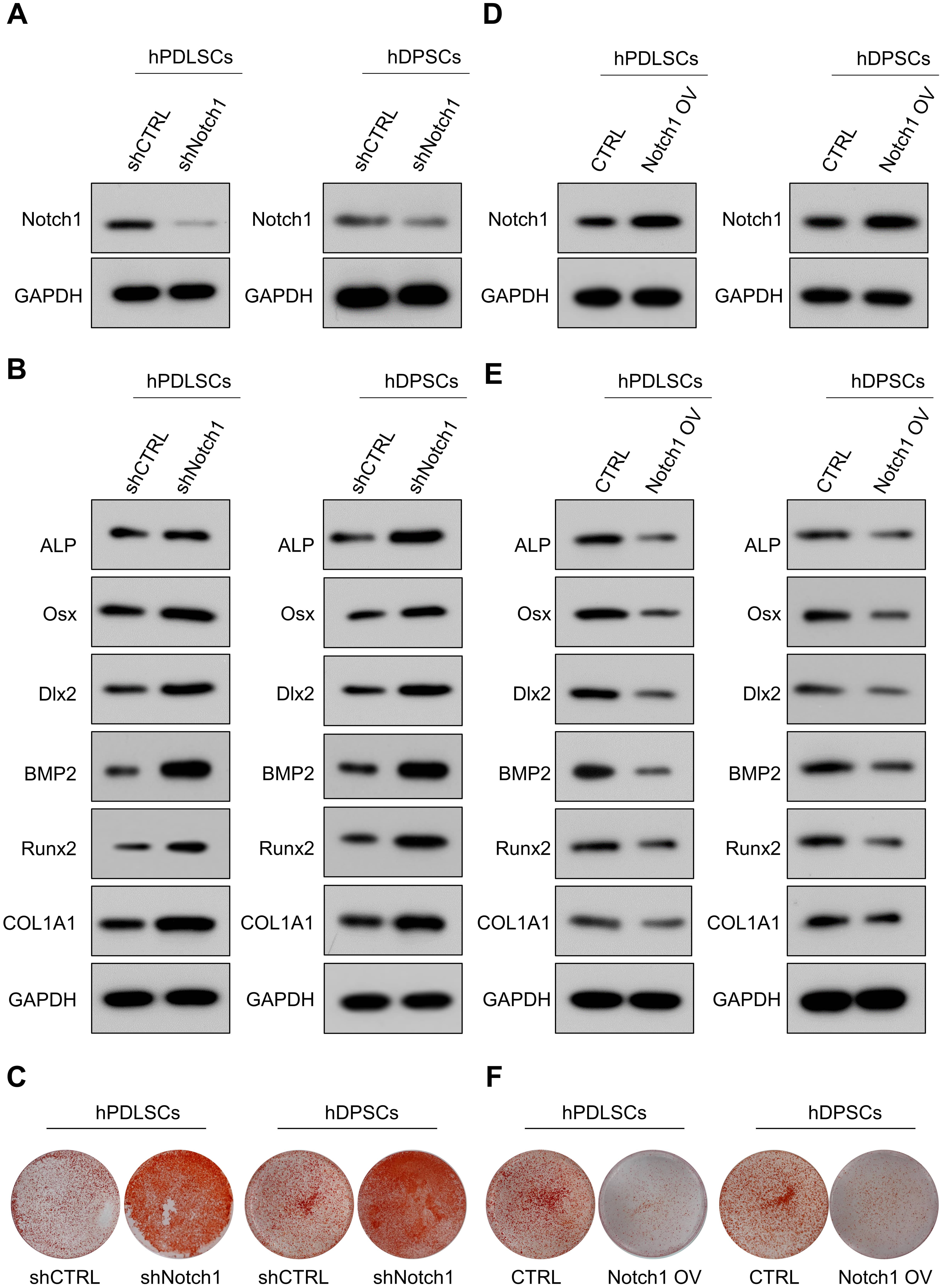 Fig. 6.
Fig. 6.Notch1 negatively regulates DSC osteogenic differentiation. (A) shRNA targeting Notch1 efficiently decreased Notch1 expression in DSCs. (B) The expression of osteogenic markers in Notch1-depleted cells and control cells. (C) The mineralized nodule-forming capacity in Notch1-depleted cells and control cells. (D) Notch1-overexpressing lentiviruses efficiently increased Notch1 expression in DSCs. (E) The expression of osteogenic markers in Notch1-overexpressing cells and control cells. (F) Mineralized nodule-forming capacity in Notch1-overexpressing cells and control cells.
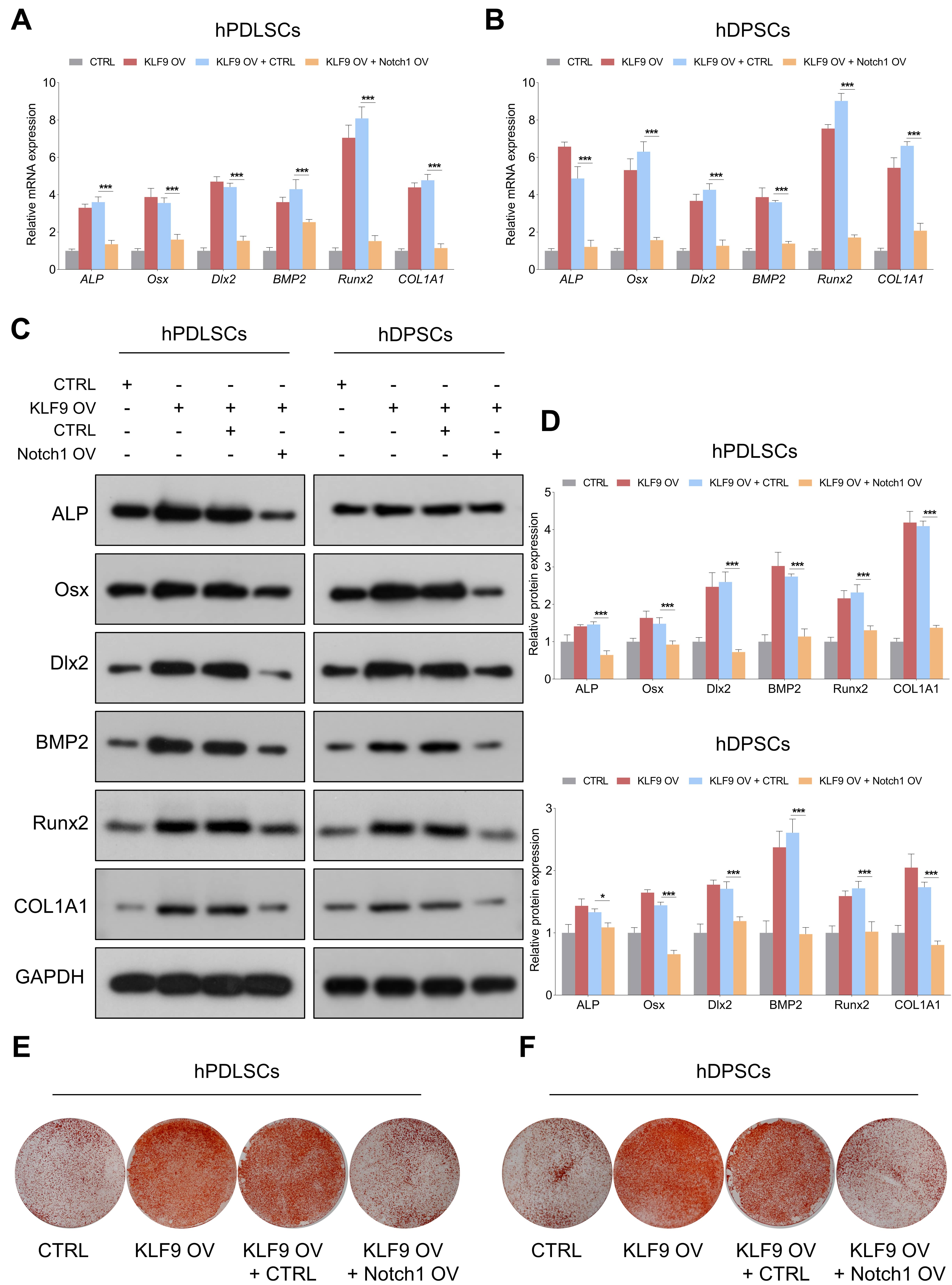
Then, we further determined whether Notch1 was a functional downstream target of KLF9 for modulating DSC osteogenic differentiation. The expression of osteogenic markers was markedly reduced in the KLF9-depleted cells, and knockdown of Notch1 almost completely reversed the suppressive effects of KLF9 depletion on the levels of osteogenic markers in DSCs (Fig. 7A–D). Similarly, the KLF9-depleted cells had significantly lower mineralized nodule formation than the control cells, and Notch1 inhibition partially rescued the mineralized nodule-forming capability of the DSCs subjected to KLF9 depletion (Fig. 7E,F).
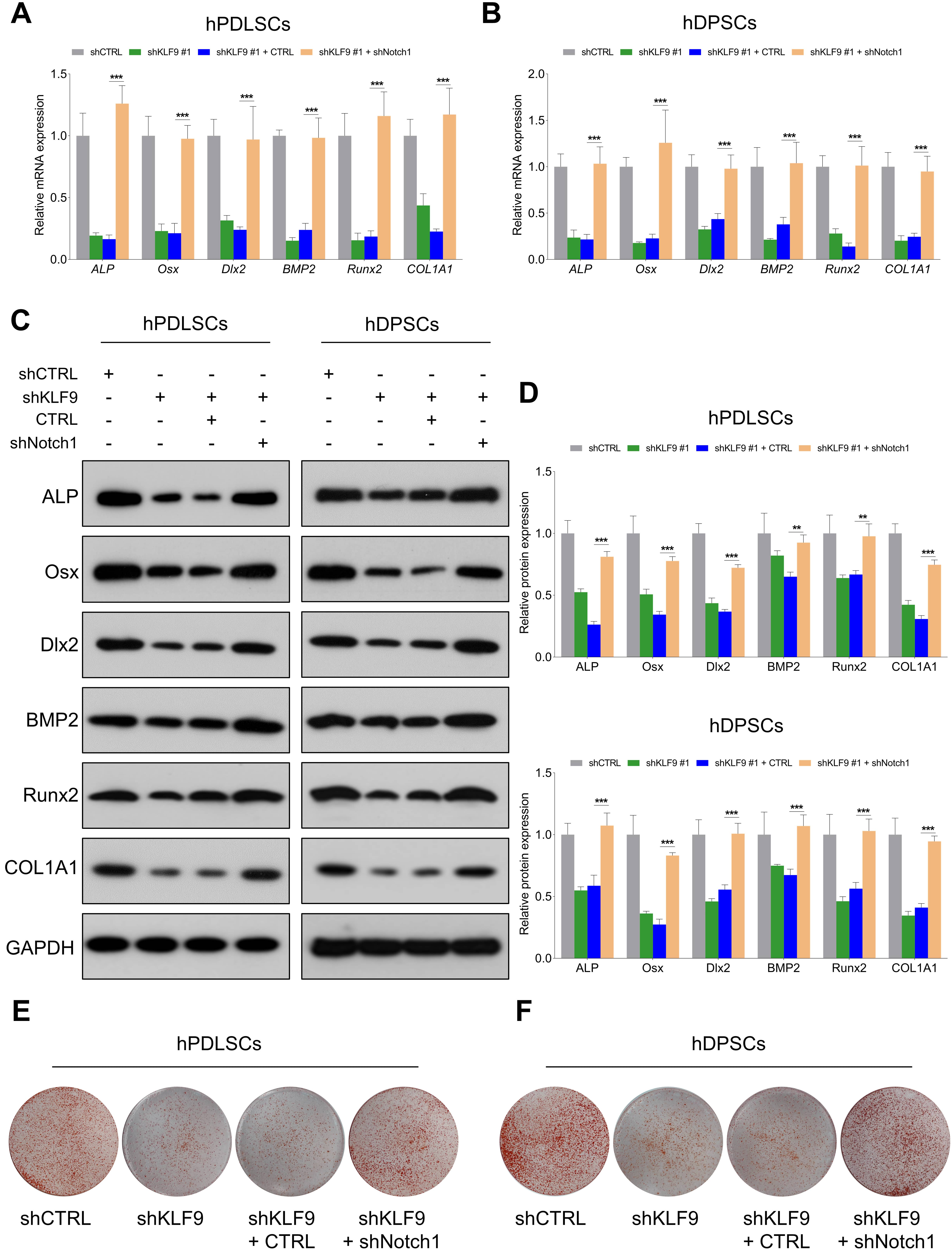 Fig. 7.
Fig. 7.Notch1 depletion partially counteracts the suppressive effects
of KLF9 depletion on DSC osteogenic differentiation. (A,B) The mRNA levels of
osteogenic markers in DSCs with the indicated modifications. (C,D) The protein
levels of osteogenic markers in DSCs with the indicated treatments. (E,F) The
mineralized nodule-forming capacity in DSCs with the indicated
treatments. **p
The effects of Notch1 overexpression on the osteogenic differentiation of KLF9-overexpressing cells were further determined. Our results demonstrated that the levels of osteogenic markers were dramatically increased in the KLF9-overexpressing cells, and enforced expression of Notch1 partially reduced the expression of osteogenic markers in KLF9-overexpressing cells (Fig. 8A–D). Similarly, ectopic expression of KLF9 markedly increased the mineralized nodule-forming capability of DSCs, and Notch1 overexpression counteracted the osteogenesis-promoting effects of KLF9 upregulation in DSCs (Fig. 8E,F).
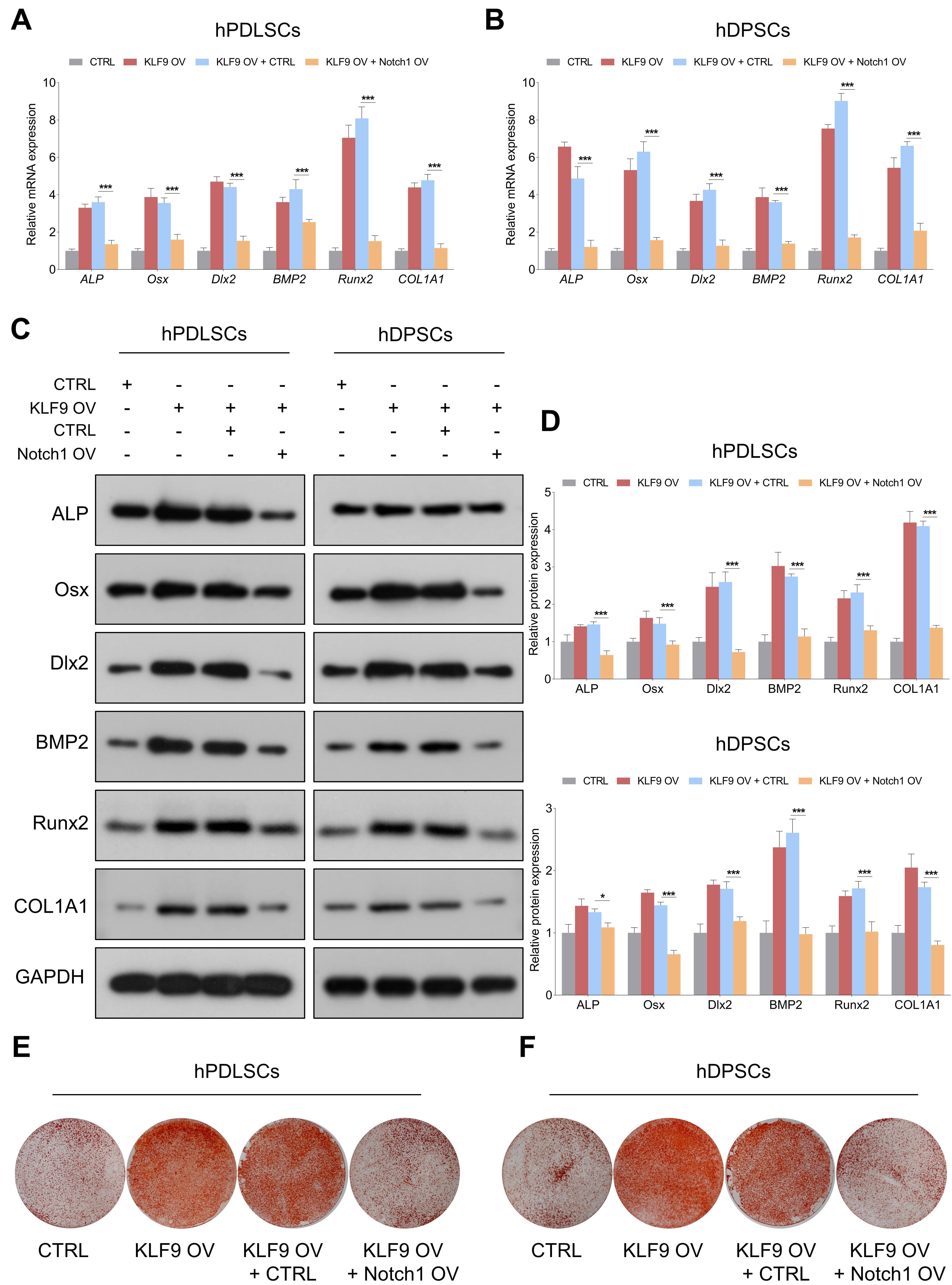 Fig. 8.
Fig. 8.Notch1 overexpression partially abrogates the enhancing effects
of KLF9 overexpression on DSC osteogenic differentiation. (A,B) The mRNA levels
of osteogenic markers in DSCs with the suggested treatments. (C,D) The protein
levels of osteogenic markers in DSCs with the indicated modifications. (E,F) The
mineralized nodule-forming capacity in DSCs transfected with the indicated
vectors. *p
In this study, we demonstrate that KLF9 is significantly upregulated during osteogenic differentiation of DSCs and other types of MSCs, suggesting that KLF9 might play an important role in modulating the osteogenic process. Our results further reveal that KLF9 depletion promotes the proliferation and suppresses the osteogenic differentiation of DSCs, and ectopic expression of KLF9 enhances DSC osteogenic differentiation. Mechanistically, KLF9 directly binds to the promoter of Notch1 and negatively regulates the expression of Notch1 and its downstream targets. More importantly, Notch1 depletion partially counteracted the inhibitory effects of KLF9 depletion on the osteogenic differentiation of DSCs, and enforced expression of Notch1 reversed the enhancing effects of KLF9 overexpression on the osteogenic differentiation of DSCs. These findings strongly demonstrate that Notch1 is a functional downstream target of KLF9.
Although KLF9 has been confirmed to be crucial for modulating many important biological processes [23, 24], its role in osteogenic differentiation remains unknown. To the best of our knowledge, we are the first to provide evidence showing that KLF9 positively regulates the osteogenic differentiation of DSCs while negatively modulating DSC proliferation. KLF9 might be an important regulator for switching cellular proliferation and differentiation in DSCs. Cell proliferation and differentiation are two distinct and mutually exclusive processes in most cells. For instance, we previously revealed that ITGA5 depletion suppressed proliferation and promoted osteogenic differentiation in DPSCs [7]. DSCs are highly proliferative cells, and their proliferative capacity gradually decreases during the osteogenic differentiation process. KLF9 may be an important transcription factor for the transition of DSCs from a proliferative state to a state of osteogenic differentiation. Upregulation of KLF9 might enable DSCs to exit the cell cycle and initiate osteogenic differentiation.
Our results showed that Notch1 negatively regulated the osteogenic differentiation of DSCs. Consistent with our findings, overexpression of the Notch intracellular domain inhibited osteogenic differentiation [25]. Similarly, Notch1 signaling was found to be suppressed by Runx2 during osteogenic differentiation [26]. Our unbiased RNA-seq data also indicated that the expression of Notch1 was reduced during osteogenic differentiation of DSCs (data not shown). However, the effect of Notch signaling on osteogenic differentiation is highly controversial, and stimulatory effects of Notch1 on osteoblast differentiation and function have also been reported [27, 28]. For instance, activation of Notch1 signaling enhances osteogenic differentiation while suppressing adipogenic differentiation in hBMMSCs [27]. We speculate that the detailed role of Notch1 signaling in mediating osteogenic differentiation might be cell type dependent and associated with the in vitro models used.
Mechanistically, we found that KLF9 directly bound to the promoter region of the Notch1 gene and negatively regulated the expression of Notch1 signaling in DSCs. More importantly, Notch1 was found to be a functional downstream target of KLF9 in modulating the osteogenic differentiation of DSCs. Although our results showed that KLF9 suppressed Notch1-mediated signaling in DSCs, there might be other important downstream targets or pathways accounting for the osteogenesis-promoting effect of KLF9 in DSCs. Therefore, omics-based technologies are needed to further determine and enrich the KLF9-mediated molecular networks in promoting DSC osteogenic differentiation.
In summary, we have demonstrated that KLF9 enhances the osteogenic differentiation of DSCs by negatively regulating the Notch1-mediated signaling pathway. These findings suggest that KLF9 plays an important role in maintaining the biological function of DSCs and provide a novel target for promoting DSC osteogenic differentiation.
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
JY and LC designed the research study. XZ, ZM, YL, and LC performed the research. XZ, ZM, YL, and LC analyzed the data. XZ, ZM, YL, LC and JY wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
This study was approved by the Ethics Committee of Nanjing Medical University (PJ2019-097-001).
Not applicable.
This work was supported by National Natural Science Foundation of China (No. 81873707, No. 82170940, No. 81901006).
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbl2805085.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.