Academic Editor: Jen-Tsung Chen
Background: The plant hormones auxin affects most aspects of plant growth and development. The auxin transport and signaling are regulated by different factors that modulate plant morphogenesis and respond to external environments. The modulation of gene expression by Auxin Response Factors (ARFs) and inhibiting Auxin/Indole-3-Acetic Acid (Aux/IAA) proteins are involved in auxin signaling pathways. These components are encoded by gene families with numerous members in most flowering plants. Methods: However, there is no genome-wide analysis of the expression profile and the structural and functional properties of the ARF and Aux/IAA gene families in soybean. Using various online tools to acquire of genomic and expression data, and analyzing them to differentiate the selected gene family’s expression, interaction, and responses in plant growth and development. Results: Here, we discovered 63 GmIAAs and 51 GmARFs in a genome-wide search for soybean and analyzed the genomic, sequential and structural properties of GmARFs and GmIAAs. All of the GmARFs found have the signature B3 DNA-binding (B3) and ARF (Aux rep) domains, with only 23 possessing the C-terminal PB1 (Phox and Bem1) domain (Aux/IAA), according to domain analysis. The number of exons in GmARFs and GmIAAs genes varies from two to sixteen, indicating that the gene structure of GmARFs and GmIAAs is highly variable. Based on phylogenetic analysis, the 51 GmARFs and 63 GmIAAs were classified into I–V and I–VII groups. The expression pattern of GmARFs and GmIAAs revealed that the GmARF expression is more specific to a particular part of the plant; for example, ARF 2, 7, and 11 are highly expressed in the root. In contrast, GmIAAs expression has occurred in various parts of the plants. The interaction of ARF with functional genes showed extensive interactions with genes involved in auxin transport which helps to control plant growth and development. Furthermore, we also elaborate on the DNA-protein interaction of ARFs by identifying the residues involved in DNA recognition. Conclusions: This study will improve our understanding of the auxin signaling system and its regulatory role in plant growth and development.
Auxin is required for growth and development at all stages of the plant life cycle. Auxin signaling is involved in a wide range of biological processes, including vegetative growth and development of numerous organs, shoot apical dominance, and the formation and differentiation of plant conducting tissues such as xylem and phloem, as well as biotic and abiotic stress responses [1]. Many active synthetic auxins have been investigated in crops for their impact on gene expression and regulation. According to previous research, two types of transcription factor families control the expression of auxin response genes: the auxin response factor (ARF) family and the Aux/IAA repressor family [2]. To control the expression of their target genes or early auxin-responsive genes, ARFs recognize auxin-responsive elements in the promoter region called AuxREs, consisting of a TGTCTC motif or variations thereof, such as TGTCGG, TCTCCC, and TCTCAC. The ARF protein has three domains: a DNA-binding domain, which incorporates the B3-like DNA binding domain (DBD); a repression or activation domain, named the middle region (MR); and a carboxyl-terminal oligomerization domain (CTD), which mediates ARF homo- and hetero-oligomerization or ARF hetero-oligomerization with Aux/IAA proteins [3, 4]. However, some genes, such as ARF13 and ARF17 in Arabidopsis thaliana, lack some domains, suggesting that each domain functions independently or that these genes are pseudo ARFs without function in the ARF-dependent pathway. The amino acid sequences of the ARFs, particularly those of the MR, determine whether they activate or inhibit the expression of auxin-responsive genes. Serine, leucine, and glutamine residues abound in the MR activation domains, while glycine, proline, serine, and threonine abound in the repression domains [5]. Domains III/IV forms the CTD and shows homology with Aux/IAA. These form the type I/II PB1 domain, which contains positive and negative charges, allowing for head-to-tail homo and hetero-oligomerization of Aux/IAA and ARF through electrostatic interactions [6, 7, 8]. The Aux/IAA proteins are assumed to bind to ARFs and inhibit auxin-responsive gene expression in the absence of auxin. By interacting with TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX (TIR1/AFB) receptors, these proteins can be ubiquitinated and degraded by the 26S proteasome at high auxin levels [9, 10, 11]. The release of ARFs allows for the regulation of auxin-responsive gene expression [12, 13]. Auxin levels fluctuate throughout tissues and developmental phases, leading to various auxin-sensing effects. Varied TIR1/AFB-Aux/IAA protein combinations have different auxin-binding affinities.
The plant physiological process is modulated by associating auxin response elements (AuxREs) of the affected genes with ARF transcription factors. For example, the AtARF1 and AtARF2 transcriptional repressors control leaf senescence, floral organ abscission, and cell proliferation in Arabidopsis thaliana [13, 14, 15, 16]. The AtARF5 regulates embryo and leaf vascular patterning [17]; AtARF6 and AtARF8 function in female and male reproduction [18]; AtARF7 and AtARF19 function in seedlings, roots, and embryo development [19]; AtARF9 operates in suspensory cells to mediate the specification of the hypophysis [20]; and AtARF10, AtARF16, and AtARF17 operate in the negative control of seed germination and post-germination activities [21]. In some instances, the ARF gene expression is influenced by exogenous auxin signals [22, 23]. Most mutations in Aux/IAA proteins disrupt auxin signaling, causing abnormalities in various developmental processes, including embryo development, hypocotyl formation, lateral root initiation and elongation, tropisms, floral organ production, and many others [24, 25, 26].
Because of the role of ARF-Aux/IAA in various physiological and developmental processes and their potential for improving crop yields, the ARF-Aux/IAA gene families have been characterized and described in Arabidopsis thaliana and essential crops such as Zea mays, Oryza sativa, Brassica rapa, and Solanum lycopersicum [23, 27, 28]. Soybeans (Glycine max) are essential for animal and human nutrition, emphasizing dietary proteins and edible oil. There has never been a comprehensive study of the gene properties, structural variation and expression of ARF and AUX/IAA protein families in soybean, although an initial search for ARFs in Glycine max was performed [29]. Physiological, genetic, and molecular methods have provided a significant amount of new informations regarding the processes of ARF-Aux/IAA in regulating auxin signal transduction and auxin degradation. This knowledge may be applied to better understand auxin signaling’s fine-tuned developmental pathways in plants. This work identified all potential GmARFs and GmAUX/IAA on a genome-wide scale. The phylogenetic relations of GmARF-Aux/IAA with other plant species such as Arabidopsis thaliana, Oryza sativa were explored.
Furthermore, the gene structure, domain distribution, and motif type of GmARF-Aux/IAAs are studied. We also provide structural models of the GmARF-Aux/IAAs proteins. The interaction of GmARF with functional genes sheds light on GmARFs potential role in plant development and growth. Finally, we compared the GmARF-Aux/IAA gene expression profiles in different soybean tissues to provide a solid basis for further functional studies of ARF-Aux/IAA genes and auxin-mediated pathways.
The soybean cultivar, Williams82, was cultivated at Northeast Forestry University Harbin, China, in a greenhouse under (26 °C, light 14 hours/dark 10 hours) growth conditions. Different parts of the 14-day-old plant were collected, such as the meristem region, unifoliate leaves, epicotyl, hypocotyl, and roots. Each sample had three separate replicas frozen in liquid nitrogen. Total RNA was isolated with Eastep® Super (Promega Beijing) according to the manufacturer’s instructions. A Nanophotomoter Spectrophotometer was used to measure total RNA’s amount and accuracy (Implen, Munich, Germany). A. thaliana, G. max, and O. sativawhole-genome and amino acid sequences were derived from the JGI Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and Ensemble databases (http://plants.ensembl.org/index.html).
The ARF and AUX/IAA gene family information was collected using the TAIR database (https://www.arabidopsis.org/) for A. thaliana. The JGI Phytozome website was used (https://phytozome.jgi.doe.gov/pz/portal.html) to compare the homology of amino acids, candidate soybean ARF, and AUX/IAA proteins with high homologous correlation with A. thaliana ARF and AUX/IAA proteins. In addition, the ARF and AUX/IAA genes were identified and assigned specific names via the NCBI database Blastp feature. The biochemistry of each ARF and AUX/IAA protein, including the number of amino acids, isoelectric point (pI), and molecular weight (MW) parameter, was determined using the online ExPASy software (https://www.expasy.org/) [30].
The amino acid sequences of ARFs and AUX/IAA proteins from G. max, A. thaliana, and O. sativa were selected to test the most suitable model using the “find best DNA/protein models (ML)” tool in MEGA 10.0 software (https://www.megasoftware.net/) to construct a maximum likelihood model. The robustness of each tree node was calculated using 1000 bootstrap replicates, with default remaining parameters [31].
To illustrate the exon-intron structure map and coding sequences (CDS) structure of GmARF-IAAs genes, the GENE Structure Display (GSDS 2.0) website (http://gsds.gao-lab.org/) was used to upload the genomic sequence and coding sequences with FASTA format to generate the gene structure. The MEME website (http://meme-suite.org/) was used to analyzed the soybean ARF and AUX/IAA proteins conserved motif using the classic mode to discover motifs on protein datasets. The generated preserved motif files are visualized with TBtools software (version 1.055) [32]. The TBtools Batch SMART plug-in links to the SMART website (http://smart.embl-heidelberg.de/), was used to determine whether there was an ARF and AUX/IAA domain in the candidate genes ARF and AUX/IAA [33].
The chromosome mapping data was acquired from the NCBI database of ARFs and AUX/IAA gene families. The chromosome mapping map was drawn with Map chart software [34]. The collinear relationship of the gene families of ARF-AUX/IAA was analyzed using MCScanx and TBtools software to further elaborate and identify orthologous and paralogous genes in the replication events of the ARFs and AUX/IAA gene families.
The 2000 bp genomic sequences upstream of the transcription start site (TSS) of the ARF and AUX/IAA gene families were extracted from the NCBI database, and the PlantCARE website analyzed the cis-acting elements in the promoter region (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [35].
The expression results were obtained through high-throughput transcriptome sequencing, and the sequencing platform was Illumina Hiseq2500 from Shanghai Biozeron Biological Technology Co., Ltd. Each sample produces 6.0-Gb sequencing data with a reading length of 150 bp, sequenced by double-terminal (PE) sequencing. Post-trimming readings have been aligned with the reference genome of G. max (Phytozome v11.0, https://phytozome.jgi.doe.gov/pz/portal.html accessed on 16 July 2016). The soybean transcriptome dataset (accession number: SRP038111) was uploaded to the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information (NCBI). The data used in the chart are the TPM values commonly used in transcriptome analysis for tissue specificity analysis and the expression of ARF and AUX/IAA gene families in different regions of the plants. Mapping these RNA-seq reads was done with the default settings using TBtools (version 1.055). Using the TPM algorithm, quantification of gene expression was performed. A clustered heatmap of the resulting TPM values for ARF and AUX/IAA per tissue was generated using the TBtools heatmap function (version 1.055). TPM data Logarithm, set base = 2 and log with = 1, and rows of clustered
The interaction networks of GmARFs and GmAux/IAAs were identified based on an orthologous-based method using the AraNetV2 (http://www.inetbio.org/aranet/) and the STRING (http://string-db.org/) databases, and the predicted interaction network was displayed using Cytoscape software. To model the three-dimensional structural homology of the spatial protein model of the GmARF/IAA gene family of soybean, we utilized SWISS-MODEL (https://swissmodel.expasy.org/interactive) supplied by the web software ExPaSy [30]. The stereo-chemical quality of each gene model was further assessed using a Ramachandran plot, which calculates structural error at each residue in the protein [36].
To identify the GmARF-IAAs gene family members in the soybean genome, the A. thaliana and O. sativa ARF-IAAs gene families were used as queries. We found the homologous soybean genes of each GmARF-IAAs using BLASTP against the NCBI database; a total of 51 GmARFs, consistent with earlier reports [29], and 63 GmIAAs genes were revealed. The proteome of soybeans was also utilized to look for HMM (Hidden Markov Model) profiles. According to their position on chromosomes, the genes were named GmARF1~GmARF51 and GmIAA1~GmIAA63, respectively. Comparative analysis of the ARF-IAAs genes revealed that in A. thaliana (ARF:23, IAA:29) and O. sativa (ARF:25, IAA:31) genes are present, but the amount of ARF:51 and IAA:63 in G. max is significantly higher than that in A. thaliana and O. sativa. The open reading frame lengths of the GmARF-IAA genes ranged from 173 to 531 amino acids, with projected PI of 4.82 to 9.48 and MWs of 19.52 to 56.83 kDa for the resulting proteins. Moreover, the number of exon and protein localization is shown in Supplementary Tables 1,2.
To further examine the evolutionary relationships in the ARF-IAAs genes, the amino acid sequences of ARFs and IAAs proteins from G. max, A. thaliana, and O. sativa were utilized in phylogenetic Maximum Likelihood trees. The gene IDs and names of ARF-IAAs genes in O. sativa and A. thaliana are shown in Supplementary Tables 3,4. A total of 99 ARFs from these three species were grouped into three subgroups (I-V) based on branching characteristics and bootstrap values of phylogenetic trees, comprising 51 GmARFs, 23 AtARFs, and 25 OsARFs (Fig. 1A). A total of 123 IAAs from these three species were clustered into seven subgroups (I–VII), including 63 GmIAAs, 29 AtIAAs, and 31 OsIAAs(Fig. 1B). The analysis shows that GmARF21 is phylogenetically close to AtARF19, while GmARF15 is phylogenetically close to AtARF9, and GmIAA39, GmIAA60 are phylogenetically close to AtIAA32, AtIAA34. Furthermore, GmIAA15, GmIAA18 are phylogenetically close to AtIAA29. There may be similarities in gene structure, protein structure, and functional domain among the same group of genes ARFs, and IAAs in A. thaliana and G. max (Fig. 1). The phylogenetic study revealed homology between A. thaliana, O. sativa, and G. max; moreover, the homology is greater in ARFs than in IAAs, suggesting that ARFs have been more conserved in evolution.
 Fig. 1.
Fig. 1.Phylogenetic relationships of the ARFs-IAAs among G. max, A. thaliana, and O. sativa. (A) ARFs and (B) IAAs. The neighbor-joining (NJ) method was used to create a phylogenetic tree utilizing the entire full-length amino acid sequences of G. max (GmARF-IAAs), A. thaliana (AtARF-IAAs), and O. sativa (OsARF-IAAs) genes, aligned to MUSCLE and MEGA (v7.0). The groups I–V in GmARFs genes and I–VII in GmIAAs genes are distinguished by a different group.
To properly comprehend the structural evolution of the GmARF-IAAs, we analyzed their gene structure, including the amount of intron/exons and functional domains. We used the GENE Structure display website (http://gsds.gao-lab.org/) to draw GmARF-IAAs gene structure; GmARF36 has no UTR area, GmARF08, GmARF30, GmARF38, and GmARF51 contain only one intron, GmARF09 contains two introns (Fig. 2A), whereas GmIAA2, GmIAA6, GmIAA31, GmIAA37 and GmIAA51 contain two introns (Fig. 2B). Some structural variations exist between ARF and IAA genes; for example, IAAs have a significantly longer UTR than ARFs, and ARFs have a higher exon/intron ratio than IAAs.
 Fig. 2.
Fig. 2.The gene structure analysis of GmARF-IAA gene family. (A) GmARFs (B) GmIAAs. The figure illustrates the exon and intron structure analyses of GmARFs and GmIAAs genes generated by the GENE Structure website. The UTRs region, exons, and introns are indicated by yellow boxes, blue boxes, and black lines. The ruler at the bottom measures the length of the exon and intron.
The conserved motif analysis of GmARFs and GmIAAs proteins revealed ten conserved motifs (motif1-motif 10) in both proteins (Fig. 3A,B), whose sequences are shown on the right side of Fig. 3. Motifs 1–5 and 8–10 are common to all GmARFs, except for GmARF10, GmARF42, GmARF49: GmARF10 only contains motif 7 GmARF42, GmARF49 do not contain motifs 3 and 9 (Fig. 3A). Except for GmIAA37, all GmIAA contain motif 1, indicating that its plays an important, possibly structural, role in GmIAA. It is worth noting that GmIAA7 and GmIAA37 only have motif 5. These two genes may be redundant in function (Fig. 3B).
 Fig. 3.
Fig. 3.The Conserved motifs of GmARFs-IAAs gene family. (A) GmARFs (B) GmIAAs. Based on the protein sequences of GmARFs and GmIAAs, the conserved motifs were discovered using the MEME website. The motif length can be calculated using the ruler at the bottom—a motif distribution map of the GmARFs and GmIAAs genes created by TBtools. The ten expected motifs are represented by coloured boxes on the left, with the Motif LOGO on the right.
The SMART website analyzes the conserved domains of GmARFs and GmIAAs (Fig. 4A,B). All GmARF genes contain a Pfam: Auxin_resp domain, a conserved region in auxin-responsive transcription factors of different species, and a B3 domain (Fig. 4A). All GmIAAs genes contain the Pfam: AUX_IAA domain. Some family members are longer and contain an N terminal DNA binding domain (Fig. 4B). Nearly all genes contain a low complexity region. The number of domains is much higher in the GmARF gene family compared to the GmIAAs gene family.
 Fig. 4.
Fig. 4.Sequence analysis for domains. (A) GmARFs and (B) GmIAAs protein sequence were analyzed using the SMART website. Pfam: Auxin resp domain and B3 domain were found in all GmARFs genes. The AUX IAA domain was found in all GmIAAs genes.
To map the chromosomal positions of GmARF and IAA genes, we use the GENE Location function of the TBtools program (Version 1.082). Two genes are found on chromosomes 11, 16, and 18, respectively. The chromosome with the most genes, chromosome 13, has 14 genes. Only GmARF genes, but no IAA genes, are found on chromosomes 11, 12, 16, and 15 (Fig. 5).
 Fig. 5.
Fig. 5.Distribution of GmARFs and GmIAAs genes in G. max genome. The 51 GmARFs genes may be found on chromosomes 1–20, whereas the 63 GmIAAs genes can be found on chromosomes 1–10, 13–15, 17, 19, and 20. The chromosomal numbers are listed at the top of each vertical bar. The ruler depicts the chromosome/Mb length. The red color shown the ARF genes while the green color for IAA genes on chromosomes.
A comparative study of the ARF and Aux/IAA proteins and homologues was undertaken to investigate the potential evolution link and compare the ARF and Aux/IAA gene family collinearity among A. thaliana and soybean using the TBtools software. The results reveal 9 (17.6%) homology for GmARF and 19 (30.6%) homology for GmAux/IAA proteins between the two species. According to this analysis, most genes have collinearity, suggesting that most genes have replication events, which increase the number of genes (Fig. 6A,B). The orthologous genes of Arabidopsis and soybean were compared. It was shown that many orthologous gene pairs between the two species also suggest a high degree of similar gene sequences. The orthologous genes of Arabidopsis and soybean showed more interspecies orthology for the IAA genes than for ARF (Fig. 7A,B), which means that the ARF genes in soybean have more variation and are less conserved.
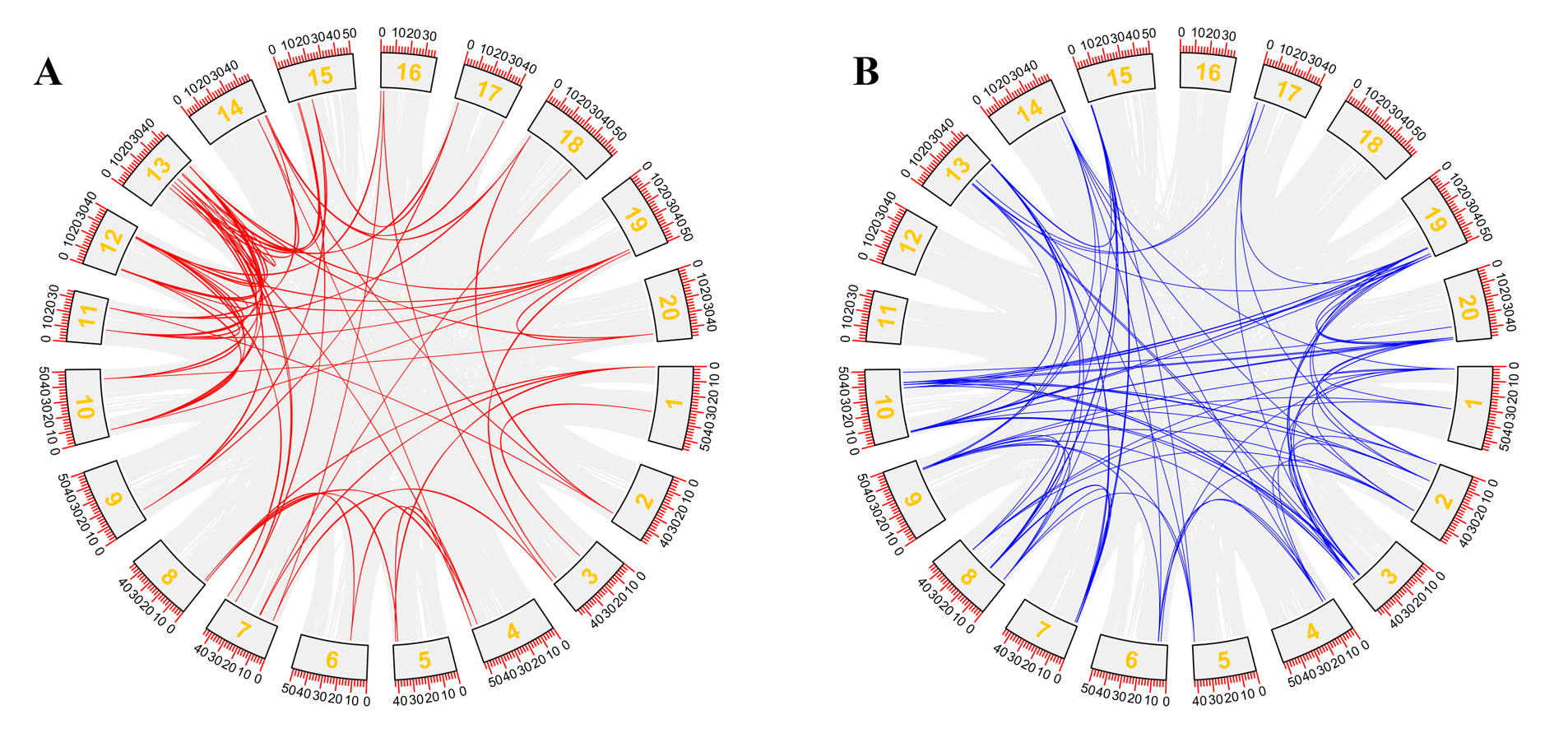 Fig. 6.
Fig. 6.Collinearity analysis of the GmARFs and GmIAAs genes. The red lines in the circle (A) are GmARFs and the blue lines in the circle (B) are GmIAAs, indicating that most of the GmARFs and GmIAAs genes have a collinear relationship.
 Fig. 7.
Fig. 7.Collinearity analysis of the GmARFs (A) and GmIAAs (B) gene families in G. max and A. thaliana. The blue lines show that most ARFs and IAAs genes had a collinear relationship between the species. Arabidopsis chromosome 1–5 is shown in pink color. The green color represents soybean chromosomes 1–20.
To elucidate the ARF, Aux/IAA gene family regulatory mechanism about plant growth and development as well as a stress response in soybean, the genomic sequence was used to query the Plant Care database to search for cis-regulatory elements (CREs). The cis-regulatory elements are the regions in the DNA that regulate the expression of the neighbouring genes. Therefore, we analyzed the cis-acting elements of each GmARF-IAAs gene in sequences located within 2 kb upstream from the transcription start site (TSS) to predict the activity of the GmARF-IAAs gene. The findings revealed various response elements, most of which were connected to various plant hormones. The promoter regions of each gene family have five hormone response elements: the IAA response element, GA response element, ABA response element, SA response element, and MeJA response element.
Furthermore, it includes elements related to response to low-temperature, defence and stress, wounds, light (MYB binding site and a part of gapA in (gapA-CMA1). The part of gapA in (gapA-CMA1) involved with light responsiveness is unique to GmARF38 and GmIAA30. Various types and numbers of cis-elements have been found in GmARFs, GmIAAs, promoters, suggesting that they participate in various regulatory processes during plant growth and development and stress responses (Fig. 8, Supplementary Table 5).
 Fig. 8.
Fig. 8.The Cis-regulatory element analysis. The cis-acting regulatory elements found in the promoter regions of GmARFs (A) and GmIAAs (B) are indicated by dots, colored according to the type of element, which are represented in the right side of the figure.
During their early stages of growth, G. max is more vulnerable to environmental stress. After 14 days of germination, total RNA was extracted using Williams82. The materials are derived from the meristem, hypocotyl, epicotyl, unifoliate leaves, and roots, among other regions of the plant. Each sample generates 6.0 GB of sequencing data, which is utilized to look into the expression patterns of the GmARF-IAAs gene families using a high-throughput transcriptome method. Fig. 9 depicts the tissue-specificity of the expression of the GmARF-IAAs genes, the corresponding FPKM (Fragments Per Kilobase per Million) values are given in the Supplementary Table 6. The heat map in Fig. 9 shows that GmARF2, GmARF5 is significantly expressed in roots in Williams82 cultivars, indicating that GmARF2, GmARF5 may conduct essential activities in roots, while the GmIAA11 is strongly expressed in the epicotyl, indicating that GmIAA11 may have essential functions in the epicotyl during the VC (Vegetative Cotyledon) stage. Furthermore, the IAAs genes family is relatively more expressed in different regions of the plant, whereas the ARFs gene family is expressed more specifically in certain organs.
 Fig. 9.
Fig. 9.Expression pattern of GmARFs-IAAs in Williams82 various tissues. (A) representing GmARFs expression, and (B) displaying the level of GmIAAs expression. The level of expression is classified in the cluster tree. M stands for meristem; U stands for unifoliate leaves; E stands for epicotyl; H stands for hypocotyl; R stands for roots. A gradual increase in value can be seen from blue to red.
Fig. 10 shows the co-expression study of GmARF-IAAs in different regions of the plant. The GmARF11,18 and GmIAA3,14,63 exhibited the greatest expression levels in the apical meristem (Fig. 10A). GmARF7,11,12,18 and GmIAA3,14,38,61 expression levels are high in true leaves (Fig. 10B). However, the expression level of GmARF genes in the epicotyl and hypocotyl are generally low, whereas high expression levels are observed for GmIAA3, 4, 11, 14, 63 in epicotyl and GmIAA1, 14, 30, 36, 39, 61, 63 in the hypocotyl (Fig. 10C,D). In roots, high expression levels are observed for GmARF2,5,11 and GmIAA9,14,63 (Fig. 10E). The expression patterns in different organs of the G. max showed that GmIAAs and GmARFs presented the highest expression levels in organ-specific, suggesting that GmARF and GmIAA control the growth and development of soybean.
 Fig. 10.
Fig. 10.Co-expression of GmARFs and GmIAAs in the same tissue of soybean. The level of expression is classified in the cluster tree. (A) M, meristem. (B) U, unifoliate leaves. (C) E, epicotyl. (D) H, hypocotyl. (E) R, roots. A gradual increase in value represents from blue to red.
To better understand the biological function and ARF and AUX/IAA regulatory networks, the protein-protein interaction (PPI) was predicted using the orthology-based method. In the absence of auxin, the AUX/IAA interacts with the ARF and inhibits its function to control the expression of auxin-responsive genes. However, when auxin was present, ARF was released from the AUX/IAA complex, performed specific activities, and modulated the expressing auxin-responsive genes. We discovered that several ARF and AUX/IAA gene families had interactions (Fig. 11A). These results confirmed the specific AUX/IAA interaction with ARF to block its function.
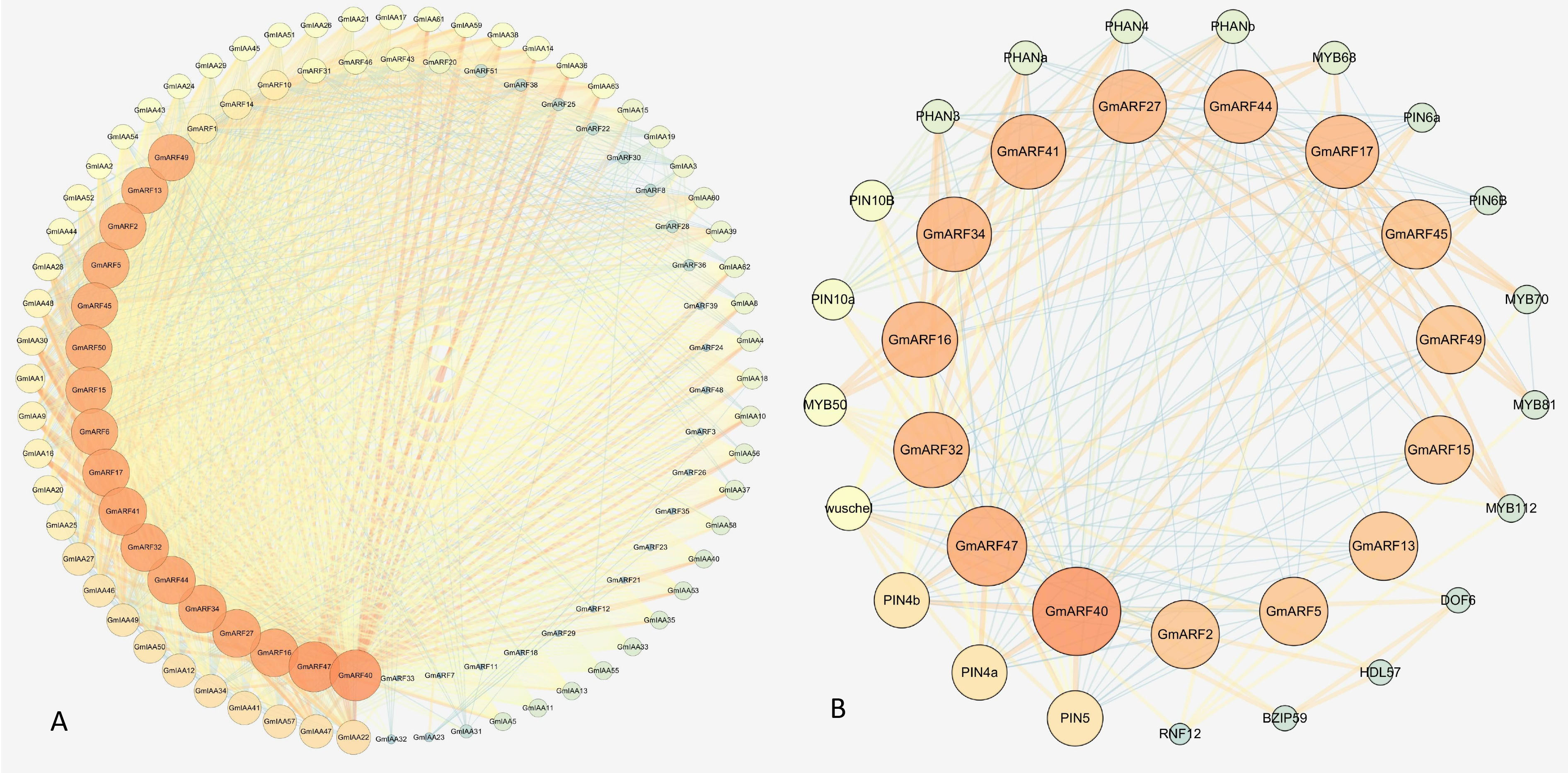 Fig. 11.
Fig. 11.Genome-wide interaction between ARF and AUX/IAA in soybean using the STRING tool. (A) The dark colour of the circle and lines represent high interaction between proteins. (B) Predicted protein-protein interaction of ARF protein with other soybean functional proteins. The inner circle represents the ARF proteins, and the outer circle represents the interacting protein. The size of the circle of each protein and dark pink colour show high interaction with other proteins.
We also analyzed the protein-protein interaction of ARF with other functional genes (Fig. 11B). As expected, most of the proteins that interacted with ARFs were important and validated components of the auxin-responsive pathways. For example, the ARF interacted with PIN (PIN-FORMED) proteins, PHAN (PHANTATUCA), MYB, and WUSCHEL; these factors played an important role in plant growth and development. A system of auxin influx and efflux transporters delivers auxin from one cell to another. PIN proteins have been postulated as auxin efflux transporters, and auxin fluxes may be anticipated based on PIN distribution in the plasma membrane, which is asymmetric in many cells. Higher plant organs have two basic axes of asymmetry: proximodistal and dorsoventral. The PHAN gene is required for dorsoventral patterns in leaves, bracts, and petal lobes. According to conditional mutants, PHAN is also essential for the early formation of the proximodistal axis. PHAN was shown to be an MYB transcription factor homolog. AUXIN RESPONSE FACTOR3 (ARF3)/ARF4 performs redundant functions in defining the abaxial leaf identity, according to independent research in Arabidopsis thaliana. A previous study found that the transcription factor MYB77 is important in Arabidopsis thaliana auxin response. As a result, Auxin-responsive gene expression was substantially reduced in MYB77 mutant plants. At low quantities of indole-3-acetic acid (IAA) and poor nutritional circumstances, the MYB77 knockout had reduced lateral root density than the wild type [37].
Homology-based 3D structure modelling was used to predict the 3D structures of
the DBDs of GmARFs and of the full length GmIAAs proteins, using the amino acid
sequences of 51 GmARF and 63 GmIAA gene family members of
soybean identified in this work. The structures were generated using the online
software Swiss-Model and revealed that the primary structures of GmARF/IAA
proteins contained both
 Fig. 12.
Fig. 12.3D structural models for GmARF and GmIAA. The GmARF/IAA protein 3D structural modelling was created using SWISS-MODEL. For the GmARFs, the DBD was modelled only. The Ramachandran plot was also used to validate the obtained models for different proteins (Supplementary Fig. 2). Further, the 3D structures of GmIAAs proteins were shown in the supplementary file.
To further explore the probable interaction profile of the GmARFs, we analyzed the homology of the residues that were reported to be important for interaction with DNA and with IAAs. The structural interfaces are shown in Fig. 13A,B [38], indicating the residues involved in the interactions. Fig. 13C,D shows the alignment of the regions of the ARF proteins that were shown to be important for DNA binding (Fig. 13C) and binding to IAA proteins (Fig. 13D) (Ref. [39]). These alignments show a high degree of homology in the residues important for the function of the ARFs, indicating that the identified GmARFs are, in fact, functional proteins.
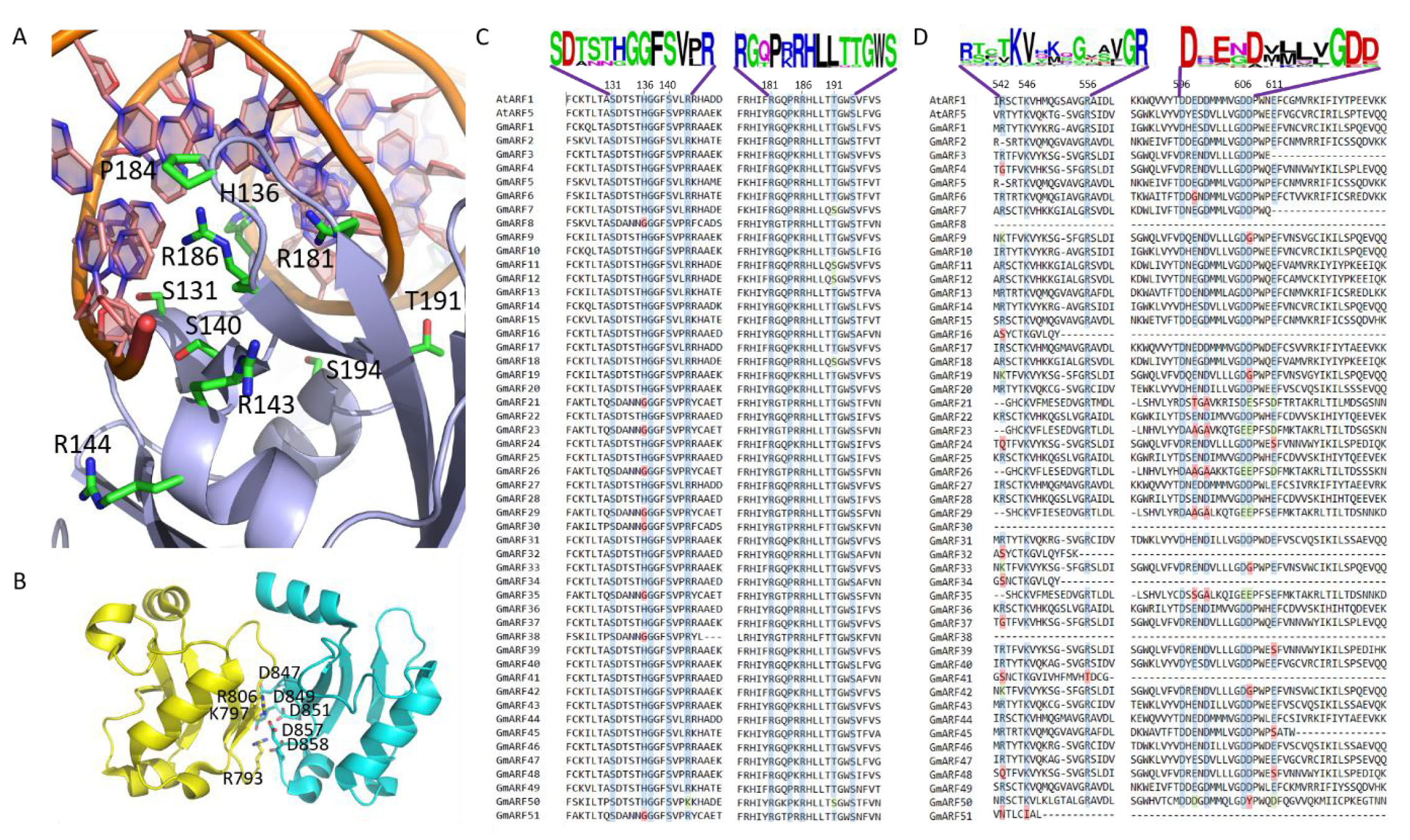 Fig. 13.
Fig. 13.Sequence alignment of important functional residues in the Arabidopsis ARF1 and ARF5, and the Glycine max ARF proteins. (A) Detail of DNA-protein interface of the ARF1-DBD/ER7 complex (PDB code 4LDX) showing the residues involved in DNA recognition. DNA-contacting residues are labelled. (B) Detail of DNA-protein interface of the ARF5-PB1 responsible for oligomerization (PDB code 4CHK) showing the residues involved in the opposite-charged patches that are important for the interaction. (C) Positions of the ARF DBD that directly contact DNA are colored, and conservation is marked using the following color codes: Blue, identical; green, conserved; yellow, semi-conserved; red, non-conserved. The residue numbers indicated above the alignment correspond to AtARF1 residues. (D) PB1 alignment: Sequence alignment of the oligomerization residues of the PB1 domains of the Arabidopsis ARF1 and ARF5, and the Glycine max ARF proteins identified in this paper. Conservation of the residues forming the charged patches, important for interactions with other ARFs and Aux/IAA proteins [39], is marked using the following color codes: Blue, identical, green, conserved; yellow, semi-conserved; red, non-conserved. The sequence logo of relevant fragments is shown at the top for the alignments shown in panels C and D.
The ARFs presumably contribute to the specificity of hormone responses by being downstream components of the auxin signaling cascade. As a result, knowing how auxin activates appropriate growth and developmental responses in a timely and tissue-specific way requires functional characterization of these transcriptional mediators. The current work provides a full picture of the soybean ARF and Aux/IAA gene family’s fundamental structural characteristics to identify the role of ARFs in mediating various auxin responses. Auxin affects cellular processes, including cell division, enlargement, and differentiation [39, 40]. Auxin early response genes, such as Aux/IAA and ARF families, are required for specific and rapid gene reprogramming in response to dynamic spatial and temporal changes in auxin levels [2]. Aux/IAA family members were reported to be short-lived nuclear proteins that suppressed the expression of ARF-activated genes. Thus, the functions of Aux/IAA are directly responsible for auxin-mediated transcriptional regulation. Aux/IAA proteins bind to ARFs and inhibit auxin-responsive genes activated without auxin [41, 42]. These proteins can be ubiquitinated and degraded through the 26S proteasome at high auxin levels by interacting with TIR1/AFB receptors [9, 10].
However, there is no genome-wide analysis of the expression profile and the structural and functional properties of the ARF and Aux/IAA gene families in soybean. We found 51 GmARF and 63 GmAux/IAA genes, almost twice as many as Arabidopsis thaliana 23 AtARF [43], 29 AtAux/IAA [44], and in rice 25 ARF [23], 31 AUX/IAA [45]. This suggests that the ARF and AUX/IAA gene families have undergone considerable duplication and diversification in soybean. It was revealed that the ARF gene family in soybean showed less homology 9 (17.6%) with Arabidopsis thaliana than Aux/IAA 19 (30.6%).
According to the analysis of conserved motifs, all GmARFs had a typical DBD domain necessary for efficient binding to AuxRE and an Auxin resp domain. Only 33 of the 53 GmARFs have the AUX IAA domain, which can mediate ARF interactions and ARF and Aux/IAA protein interactions. Since the AUX IAA domain is required for multimerization, it is therefore important to investigate how these ARFs act and whether they require interactions with other proteins. For those ARFs that incorporate a DBD and an AUX/IAA domain, we find that the residues important for DNA and IAA binding, respectively, are highly conserved, suggesting conservation of the functionality of these interactions.
Further, the analysis of the gene structure showed that the number of exons in the GmARF genes ranged from 2 to 14, and in GmIAA ranged from 3–6, while similar results were found in Arabidopsis [22], rice [23], suggesting that the family of plant ARF genes has highly conserved structures and potentially similar roles across dicotyledonous and monocotyledonous plant species. The number and size of the introns between ARF and AUX/IAA gene family were significantly high; the Aux/IAA gene family have a very high length of the intron, which is a non-coding part of the genes. According to previous literature, the size and number of introns have an important evolutionary role and modulate different regulatory functions in gene expression [46, 47].
The GmARF and AUX/IAA gene expression profiles provide insights into potential functions for these genes. The majority of GmARFs are concentrated in the meristem regions of plants, with a low GmARF expression in the soybean epicotyl. An opposite pattern is observed for the GmIAAs, which show high expression levels of expression in the epicotyl and low levels in the meristem. Furthermore, certain ARF and AUX/IAA demonstrated specific expression patterns in different plant tissue [48]. Many research groups have confirmed the expression pattern and its developmental role of ARFs. In Arabidopsis and other plant species many candidate genes have been proposed to be controlled by auxins that may act in growth and developmental processes [7, 49]. The ARF family members have been suggested to have a crucial role in regulating the expression of auxin response genes [24]. ARF genes express dynamic and differential patterning during development, and genetic investigations have revealed that unique ARF influence various developmental processes [20]. Members of the ARF family of proteins comprise domains involved in DNA binding, transcriptional activation or repression, and protein-protein interactions important for auxin perception and signaling [50]. Our analysis from the STRING database confirmed that ARF gene family members have direct interaction with different genes, which regulate different aspects during plant growth and regulation. Through their DNA-binding domain, ARFs directly bind to AuxREs (Auxin Responsive Elements) in the promoters of auxin-responsive genes [51]. The C-terminal amino acids are also required for ARF binding to AuxREs [2]. The C-terminal domain is thought to increase DNA binding by allowing ARF oligomerization. Domains III and IV are conserved regions at the C-terminus of ARF and Aux/IAA proteins [2]. According to yeast two-hybrid and bimolecular fluorescence complementation tests, these domains facilitate ARF-ARF, ARF-Aux/IAA, and Aux/IAA-Aux/IAA interactions [19]. The C-terminal domains of ARF5 and ARF7 were found to correspond to a well-known PB1 domain, which confers protein-protein interactions with other PB1 domain proteins through electrostatic contacts, as shown by their crystal structures [19, 38]. The importance of these charged amino acids in conferring ARF and Aux/IAA interactions, as suggested by the crystal structure of the PB1 domain, was confirmed in more experiments [19]. Structure-function analysis and saturating binding site selection on the ARF1 and ARF5 DNA binding domains have recently revealed a second protein-protein interaction module that functions in ARF-ARF dimerization [4]. ARFs have been shown to regulate and be regulated by other transcription factors.
In plants, the PIN-FORMED (PIN) proteins regulate the auxin distribution and concentration gradients in various tissue [52, 53]. Auxin treatments induce the expression of PIN1, PIN3, and PIN7, particularly in roots [54, 55, 56, 57]; during organogenesis, the action of the PIN proteins resulted in spatiotemporal auxin maxima that trigger the establishment of new growth axes. Members of the HD-Zip superfamily, AP2/EREBP-type transcription factors, AS2-like (LBD) and MYB-like transcription factors, zinc finger-like transcription factors, and others are all known to be auxin-induced transcription factors [58, 59, 60, 61], but the functions of these elements in certain developmental stages are poorly understood in general. In conclusion, auxin signal attenuation requires the PB1 domains of the ARF and Aux/IAA proteins. The future analysis of the ARF and Aux/IAA protein PB1 interface may provide insight into the ARF and Aux/IAA protein complexes’ in planta specificities.
In the present study, we identified 53 ARF and 63 AUX-IAA genes in the soybean genome. We analyzed their phylogenetic, gene structure, conserved domain, and motifs to determine their evolutionary relationship with other plant species. The expression profiles in various organs uncovered the expression diversity of ARF and IAA genes family in soybean. Furthermore, the analysis of the interactions of the GmARF with different functional genes demonstrates its role in plant growth and development. These findings will serve as a strong foundation for future research into the functional characterization of ARF, AUX-IAA genes, and ARF-mediated signal transduction pathways, enabling us to learn more about the molecular basis of soybean genetic enhancements. In addition, because the auxin-mediated signal transduction pathway is complex, more systematic research on ARF’s auxin signaling pathway target genes, such as SAUR and GH3, is needed better to understand soybean development, ripening, and environmental adaptation.
Conceptualization—SA; Methodology and writing—original draft preparation—SA, WW, ZZ; Writing—review and editing—NK, DRB & LX; Data curation—SA, WW. All authors have read and agreed to the published version of the manuscript.
Not applicable.
Not applicable.
This work was supported by Ministry of Economy and Competitiveness of the Spanish Government grants PID2020-117028 GB-I00 (AEI/FEDER, EU), BIO2016-77883-C2-2-P (AEI/FEDER, EU) and FIS2015-72574-EXP (AEI/FEDER, EU) to D.R.B.
The authors declare no conflict of interest. NK is serving as one of the Editorial Board member and Guest Editor of this journal. We declare that NK had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to JTC.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
