Academic Editor: Josef Jampílek
Background: Avascular necrosis of the femoral head (AVNFH) is a progressive, multifactorial, and challenging clinical disease that causes hip pain and loss of hip joint function. Till now, the pathogenesis of AVNFH was not fully understood. In this study, we characterized cartilage protein profiles of patients with AVNFH and identified the potential proteins involved in the progress of AVNFH using proteomics technique. Methods: Proteins from the cartilage of 6 patients (3 AVNFH patients and 3 fracture patients) were extracted and identified using label-free proteomics. AVNFH-responsive proteins were compared with those of the fracture patients and duly identified. Bioinformatics analyses including gene ontology (GO), KEGG, and STRING were performed to identify the functions of AVNFH-responsive proteins. Results: A total of 1512 proteins were identified from cartilage tissues of the patients. Compared to fracture patients, 255 significantly changed proteins were identified in cartilage tissues of patients with AVNFH. Functional categorization indicated that the significantly changed proteins were mainly involved in ECM-receptor interaction, focal adhesion, and glycolysis pathways. Interestingly, adipocyte enhancer-binding protein 1, cytoskeleton-associated protein 4, and ASPN protein were dramatically decreased, however, anti leukoproteinase, erythrocyte membrane protein, and lysozyme c were highly increased in patients with AVNFH. Conclusions: The current proteomic results suggest that ECM-receptor interaction and focal adhesion related proteins contribute to development of AVNFH. To our knowledge, this is firstly reported proteomic study on cartilage tissues of patients with AVNFH. The marker proteins including caveolae-associated protein 3 and procollagen-lysine 2-oxoglutarate 5-dioxygenase 2 could help us to understand the pathogenesis of AVNFH.
Avascular necrosis of femoral head (AVNFH) is a progressive, multifactorial and challenging clinical disease that commonly affects 30- to 50-year-old individuals [1]. Once AVNFH occurs, patients suffer from hip pain and loss of the hip joint function [1, 2, 3]. It is a common and frequently-occurring clinical disease in orthopedics. Currently, there are more than 20 million patients in the world. Recently, with the indiscriminate use of hormonal drugs [4], increasing number of people who drink alcohol [5], and the increase of hip high-energy trauma (high-speed traffic accident injury, high fall injury of construction workers, high injury rate of sportive people, etc.) [6], the incidence of AVNFH seems to increase more rapidly than other musculoskeletal diseases. Pathological features of AVNFH comprise necrosis of the femoral head, collapse and secondary osteoarthritis of the hip joint [7], which lead to intractable pain, claudication and limb length differences in patients, ultimately resulting in high disability rates. Till now, the pathogenesis of AVNFH was not fully understood.
The common known treatments for AVNFH include microwave therapy, magnetic therapy, physical therapy, far-infrared drilling decompression, and bone flap transplantation surgery [8]. Most of these known treatment methods are confined to the symptomatic management of AVNFH and are unable to block or reverse the underlying ischemic hypoxia of the pathological processes of necrotic bone cells. Patients eventually choose femoral head resection or artificial joint replacement. These are not only time-consuming processes but are usually accompanied by pain and even mental trauma. Conventional magnetic resonance imaging (MRI) is useful for diagnosing AVNFH [9, 10], but it is not sensitive to biochemical changes which precede the morphological changes of AVNFH. Proteomics is defined as the high-throughput study of proteins [11, 12]. Proteomics via state-of-the-art quantitative techniques and bioinformatics enables identification, quantitation, validation and characterization of a variety of proteins from specific organs, tissues or cells. The information obtained therein aside from aiding to determine protein structure, underlying enzymatic mechanisms and regulatory functions, provide invaluable links between protein levels and diseases.
In this study, a comparative clinical proteomic analysis was performed to profile the proteome of patients with AVNFH. Through identification of AVNFH-marked proteins, we sought to at least in part, uncover the pathogenesis of AVNFH and contribute to determining potential novel targets for clinical intervention.
The review board of the First Affiliated Hospital of USTC, University of Science and Technology of China approved this study. Six fragments of articular cartilages from 6 patients (3 men, 3 women; mean age, 61.8 years) (Table 1) with AVNFH and fracture were used for this study. AVNFH was diagnosed based on its established diagnostic criteria [13, 14].
| ID | Age | gender | Height (cm) | weight (kg) | BMI (kg/m |
Pathological feature |
| Patient 1 | 52 | male | 170 | 65 | 22.49 | femoral head necrosis |
| Patient 2 | 54 | female | 165 | 60 | 22.04 | femoral head necrosis |
| Patient 3 | 58 | female | 155 | 68 | 28.30 | femoral head necrosis |
| Patient 4 | 78 | female | 163 | 55 | 20.70 | fracture |
| Patient 5 | 62 | male | 172 | 70 | 23.66 | fracture |
| Patient 6 | 67 | male | 168 | 72 | 25.51 | fracture |
| BMI, Body mass index. | ||||||
For extraction of proteins from the articular cartilages, the
Tris–phenol and Acetone precipitation methods were used. Details of the
Tris–phenol method is as follows: a portion (0.1 g) of each sample was ground to
powder in liquid nitrogen using a mortar and pestle and then mixed with 1 mL
extraction buffer (500 mM Tris-HCl, 50 mM EDTA, 700 mM sucrose, 100 mM KCl, 2%
The acetone precipitation method was performed as follows: the sample was ground in liquid nitrogen with a mortar and pestle and then mixed with extraction buffer composed of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, phosphatase inhibitor mixture (Sigma, St. Louis, MO, USA), and protease inhibitor mixture (Roche, Werk Penzberg, Germany). After centrifugation at 12,000 xg for 20 min, the obtained supernatant was diluted with three volumes of cold acetone containing 0.07% 2-mercaptoethanol and then incubated at –20 °C for 2 h. The resulting precipitate was collected and washed three times with an acetone solution containing 0.07% 2-mercaptoethanol. Finally, the protein pellets were dried in a Speed-Vac and resuspended in a lysis buffer which consisted of 7 M urea, 2 M thiourea, 5% CHAPS, and 2 mM tributylphosphine. The protein concentration was then measured using the Bradford method with bovine serum albumin as the standard [15].
The purified proteins were reduced and alkylated according to the manufacturer’s
instructions (Sigma, St. Louis, MO, USA) with some modifications. Briefly, the
protein solution containing 100
Peptides in 0.1% formic acid were loaded onto an EASY-nLC1000 nano-liquid
chromatography (LC) system (Thermo Fisher Scientific, San Jose, CA, USA) equipped
with a C18 PepMap trap column (100
Raw mass spectrometry (MS/MS) spectra search was conducted using MaxQuant-associated Andromeda search engine [16], and a uniprot-Homo sapiens database (82616 sequences). Initial maximum precursor and fragment mass deviations were set to 6 ppm and 0.5 Da, respectively. Variable modification (methionine oxidation and N-terminal acetylation) and fixed modification (cysteine carbamidomethylation) were set for the search and trypsin with a maximum of two missed cleavages was chosen for searching. The minimum peptide length was set to 7 amino acids and the false discovery rate (FDR) for peptide and protein identification was set to 0.01. The precursor ion mass accuracy was improved using the time-and mass-dependent recalibration option. Frequently observed laboratory contaminants were removed and the protein identification was considered valid only when at least 2 matched peptides and 1 unique peptide were present.
The freely available software Perseus (version 1.4.1.3) (available online:
http://141.61.102.17/perseus_doku/doku.php?id=start) was used to compare the
peak intensities across the whole set of measurements to obtain quantitative data
for all of the peptides in the sample. The label-free quantitation (LFQ)
intensities of proteins from the MaxQuant analysis were imported and transformed
to logarithmic scale with base 2. The missing values were replaced with the value
of the lowest intensity. The protein quantification and calculation of
statistical significance was carried out using Student-t test and error
correction (p-value
To perform the functional analysis, the identified proteins were analyzed using QuickGO (http://www.ebi.ac.uk/QuickGO/). Pathway mapping of identified proteins was performed using the KEGG database (http://www.genome.jp/kegg/). Protein-protein interaction was analyzed using the STRING database.
To uncover the inherent mechanism of femoral head necrosis, comparative proteomics was performed using cartilage tissues from AVNFH and fracture patients (Fig. 1). The AVNFH and fracture patients were diagnosed by X-ray scans (Fig. 2). Proteins from cartilage tissues were extracted by both the Tris–phenol method and acetone precipitation method. SDS-PAGE analysis indicated that proteins extracted by Tris–phenol had more bands than those extracted by the acetone precipitation (Fig. 3). The proteins extracted using Tris–phenol method were then subjected to LC-MS/MS analysis. A total of 1510 proteins were identified from the cartilage tissues of AVNFH and fracture patients (Fig. 4A, Supplementary Table 1). Among them, 1419 proteins were common between the AVNFH and fracture patients (Fig. 4B). Also, 81 and 10 proteins were specifically identified in the AVNFH and fracture patients, respectively (Fig. 4B, Supplementary Table 2). For the common proteins, 88 were significantly increased while 74 proteins were decreased, respectively in the AVNFH patients compared to the fracture patients (Fig. 4B, Supplementary Table 3).
 Fig. 1.
Fig. 1.Experimental design of the study. Images of articular cartilages from patients with avascular necrosis of femoral head and fracture were captured using nuclear magnetic resonance (NMR). The articular cartilages were comparatively analyzed using proteomics. The functions of identified marker proteins were analyzed using Gene Ontology (GO), KEGG pathway, and STRING database.
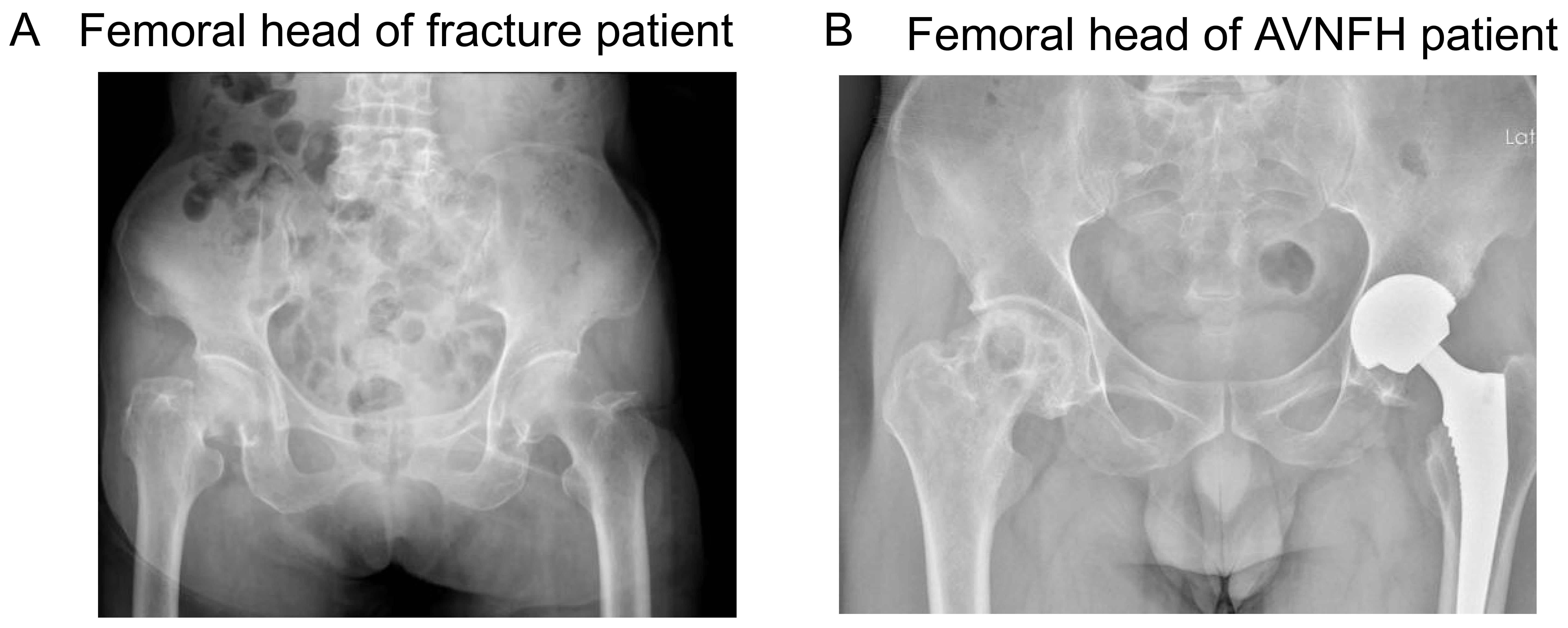 Fig. 2.
Fig. 2.X-ray images of femoral head from fracture and AVNFH patients. (A) X-ray image of femoral head from fracture patient. (B) X-ray image of AVNFH patient.
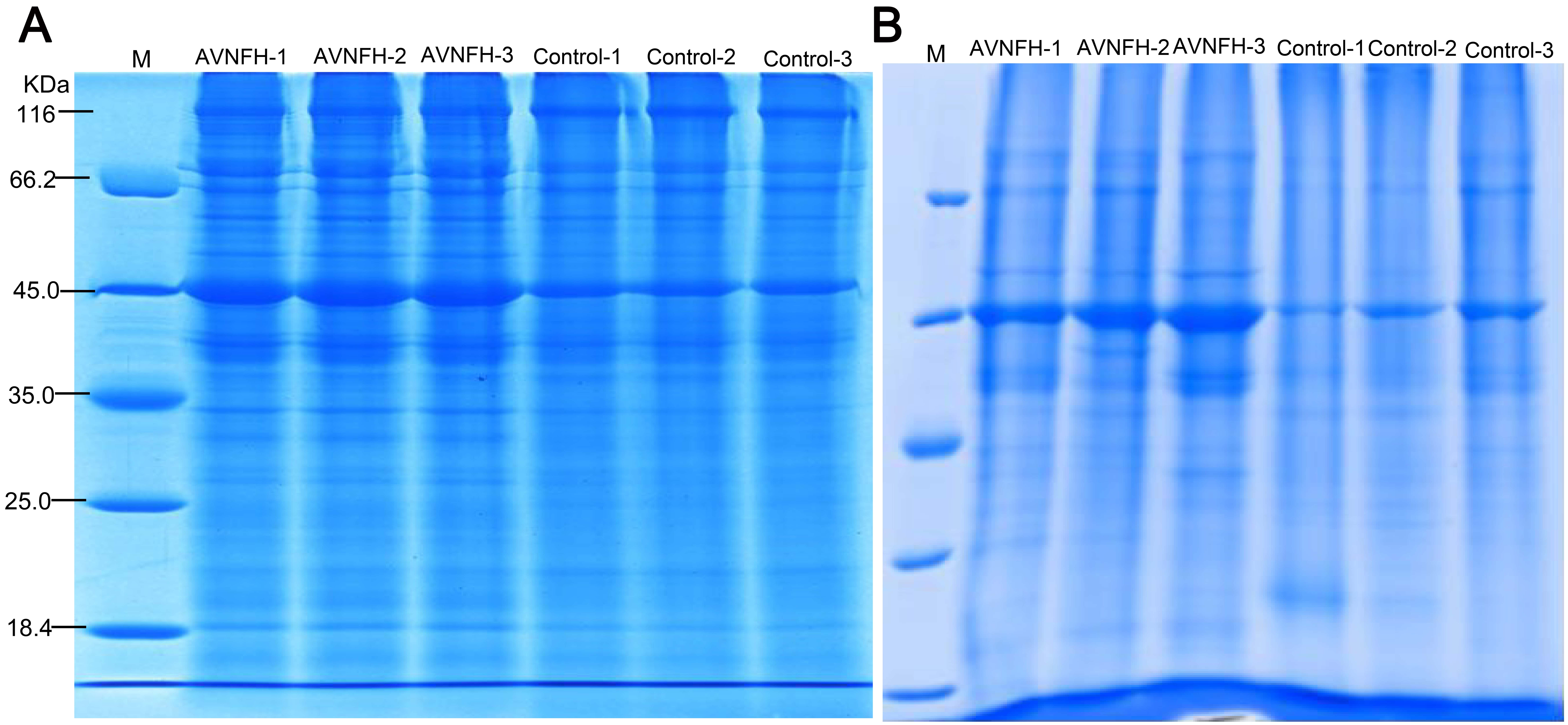 Fig. 3.
Fig. 3.Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of extracted proteins from AVNFH patients and fracture patients (control). (A) Proteins were extracted from AVNFH patients and fracture patients (control) using Tris-phenol buffer. After extraction, protein concentration was determined using BCA kit and protein quality was analyzed by SDS-PAGE. (B) Proteins were extracted from AVNFH patients and fracture patients (control) using Acetone precipitation method. After extraction, protein concentration was determined using BCA kit and protein quality was analyzed by SDS-PAGE.
 Fig. 4.
Fig. 4.Significantly changed proteins between AVNFH and fracture patients. Proteins extracted from articular cartilages were digested, analyzed using nano-LC-MS/MS, and compared through MaxQuant analysis. The comparison was performed using abundance of protein in articular cartilages of patients with AVNFH divided by those in patients with fracture. (A) Volcano plot showing the significantly changed proteins. (B) Venn diagram showing the distribution of identified proteins in fracture and AVNFH patients.
To understand the cellular processes and functions of the significantly changed proteins, GO and KEGG databases were used. Through these bioinformatic analyses, abundant common proteins (Fig. 5A) and unique proteins (Fig. 5B) in either the AVNFH patients or fracture patients were enriched in biological processes, cell components, molecular functions, and KEGG pathway. At the GO level, the significantly changed proteins were mainly involved in anatomical structure development, extracellular region/organelle, and cell adhesion molecule binding (Fig. 6A). The unique proteins in either the AVNFH or fracture patients significantly related to anatomical structure development, extracellular vesicle, and protein binding (Fig. 6B).
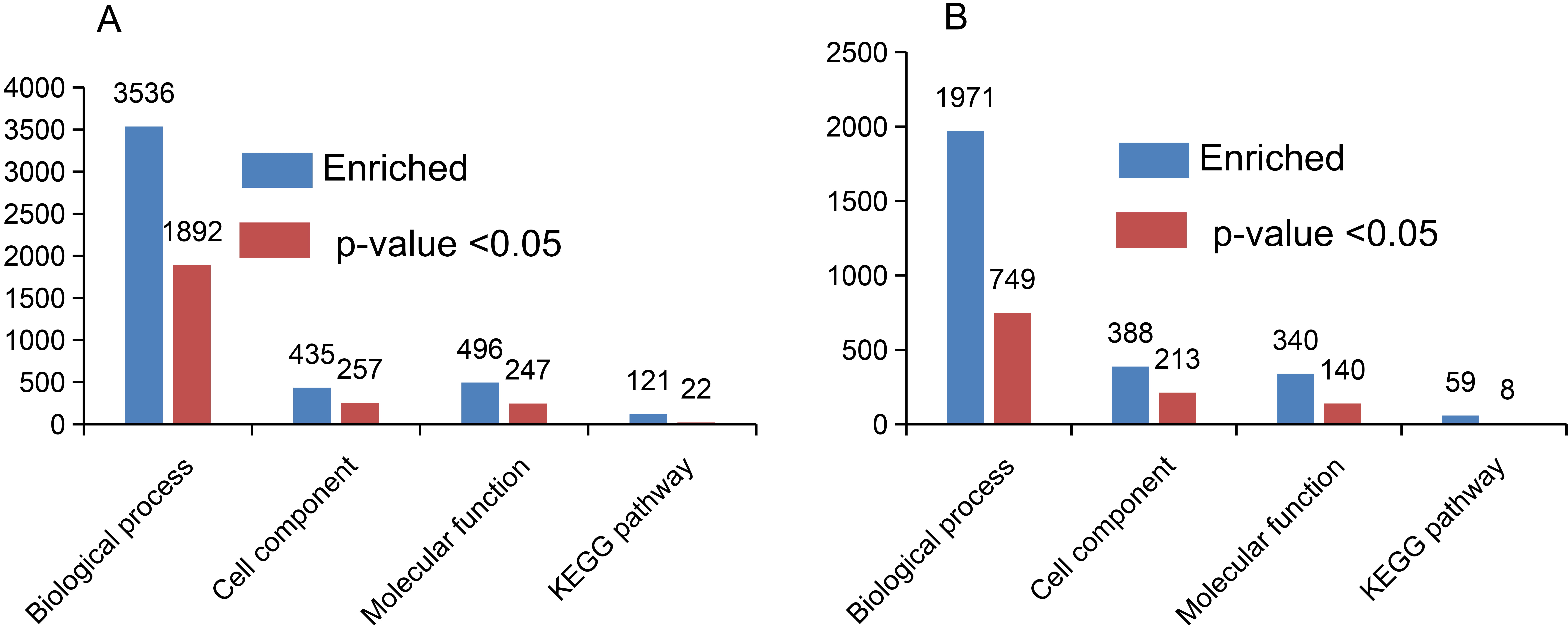 Fig. 5.
Fig. 5.Statistical analysis of GO and KEGG pathways of significantly
changed proteins. (A) The number of KEGG pathways and GO categorization
including biological process, cell component, and molecular function, the
commonly identified proteins are involved. (B) The number of KEGG pathways and GO
categorization including biological process, cell component, and molecular
function for the fracture-specific and AVNFH-specific proteins. The orange color
indicates significantly enriched proteins (p
 Fig. 6.
Fig. 6.Detailed information of functional categorization of significantly changed proteins. (A) Details of GO analysis of common proteins identified in both fracture and AVNFH patients. (B) Details of GO analysis of proteins specifically identified in only the fracture or AVNFH patients.
To further determine the biological pathways that are mediated by the identified proteins in cartilage tissue from AVNFH, the significantly changed proteins were analyzed using the KEGG database. These proteins were mainly involved in ECM-receptor interaction, focal adhesion, and complement and coagulation cascades (Fig. 7A). The unique proteins in either the AVNFH or fracture patients significantly related to complement and coagulation cascades, phagosome, and ECM-receptor interaction (Fig. 7B).
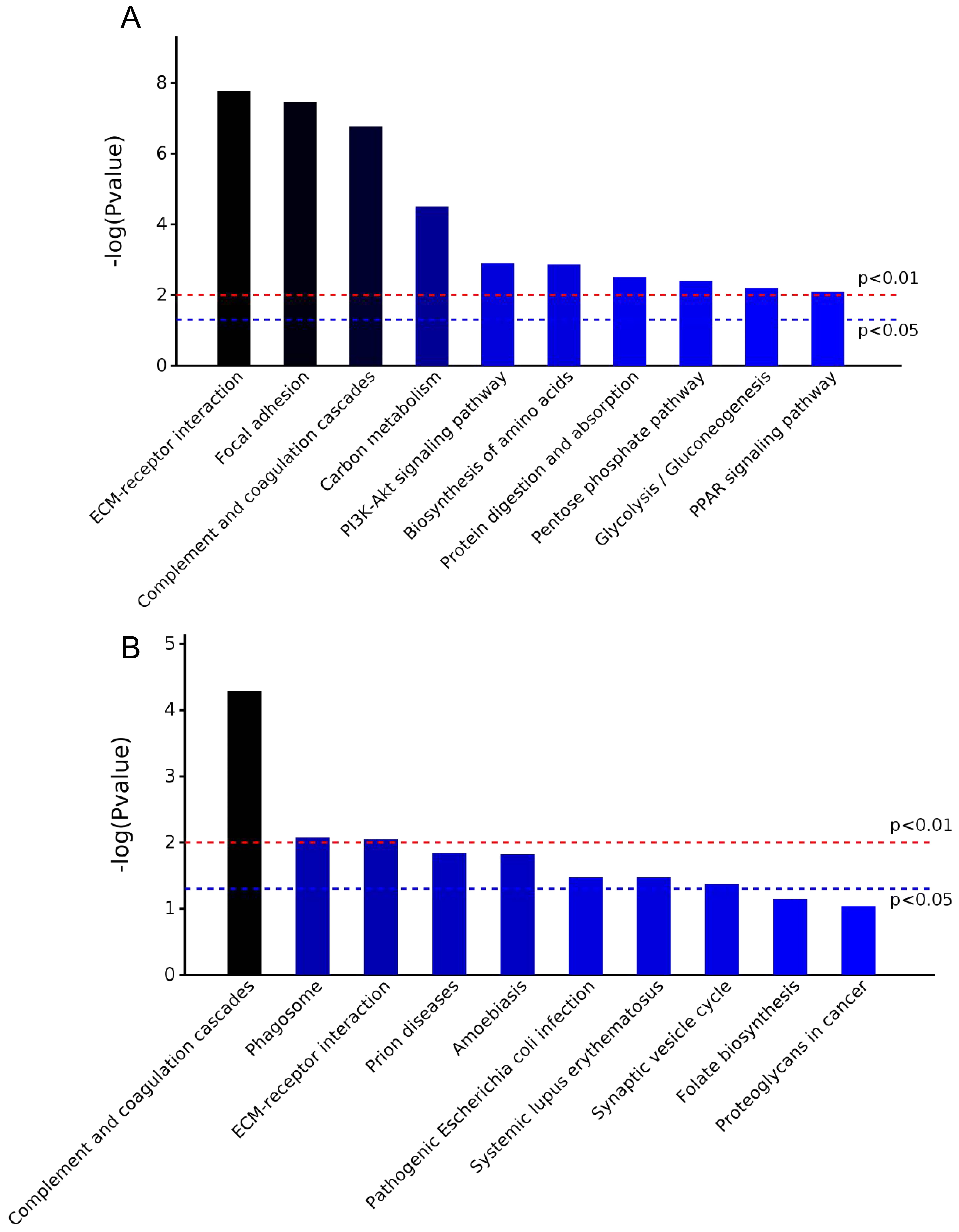 Fig. 7.
Fig. 7.Detailed information of pathway enrichment analysis. (A) Common proteins identified in both fracture and AVNFH patients were analyzed using KEGG database. (B) Pathways of proteins specifically identified in either the fracture or AVNFH patients involved were also analyzed using the KEGG database.
To determine the proteins that play central roles in pathological development of
AVNFH, protein-protein interactions of AVNFH responsive proteins were analyzed
with reference to the STRING database. As a result, increased proteins including
L-lactate dehydrogenase B chain, hemopexin, and endoplasmic reticulum lumenal
Ca
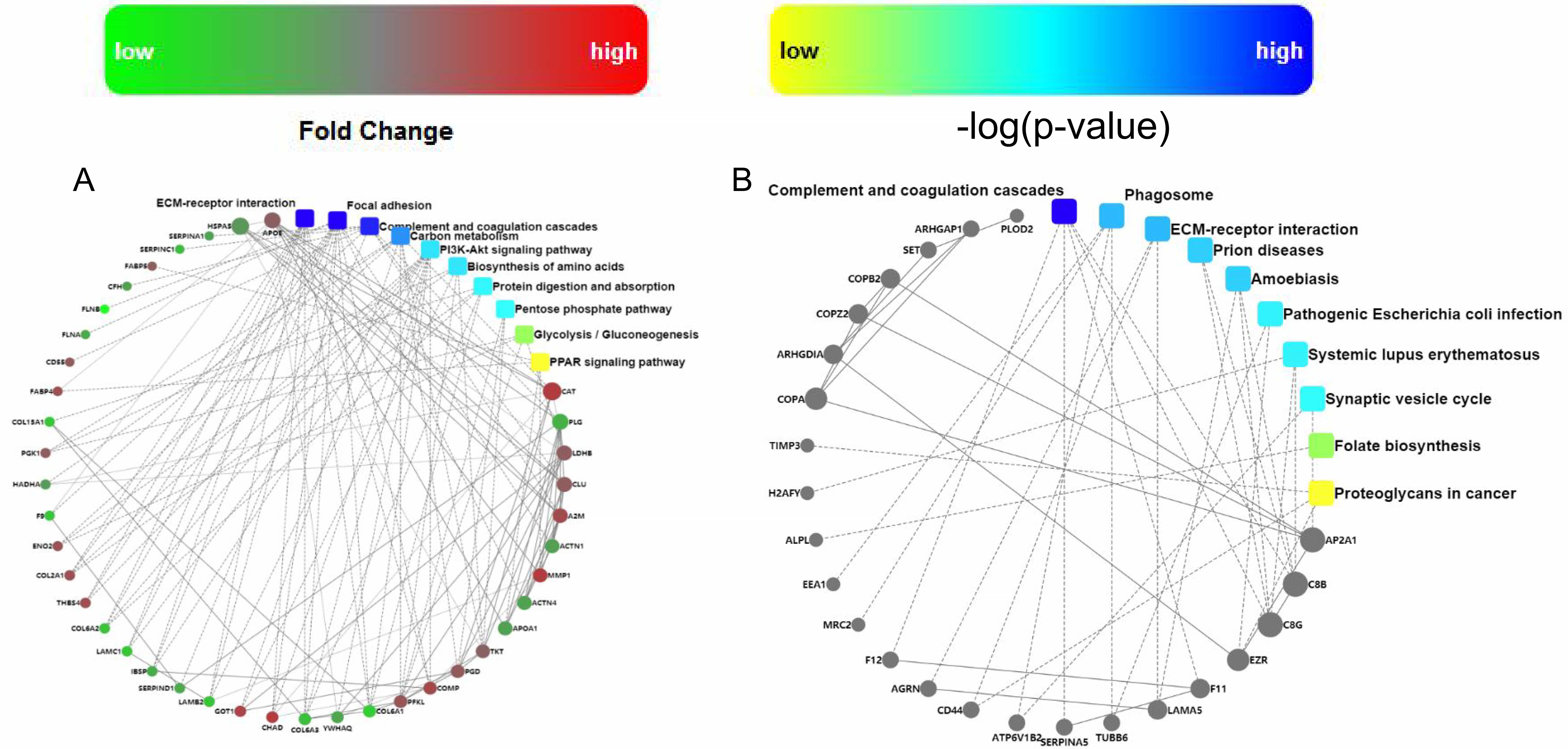 Fig. 8.
Fig. 8.Protein-protein interaction analyses of identified proteins. (A) Protein-protein interaction analysis of common proteins identified in both the fracture and AVNFH patients. (B) Protein-protein interaction analysis of proteins specifically identified in either the fracture or AVNFH patients. Colors from green to red indicate the change tendencies of proteins while colors from yellow to blue show significant values.
In our study, proteins from the cartilage tissues were extracted by Tris-phenol buffer method and acetone precipitation method. SDS-PAGE analysis indicated that the Tris-phenol extracted proteins had more bands compared to the acetone extracted proteins (Fig. 1). Protein extraction using phenol was first reported by Hurkman and Tanaka in 1986 [17]. The advantages of phenol extraction included efficient protein recovery and nonprotein components removal. The use of phenol is known to be more efficient in extracting proteins from recalcitrant tissues [18]. Protein extraction from cartilage tissues is difficult compared to protein extraction from cell. Previous protein extraction from cartilage tissues using chaotropic buffer (4 M GdnHCl, 50 mM NaAc, 100 mM 6-aminocaproic acid, 5 mM benzamidine, 5 mM N-ethylmaleimide, pH 5.8) identified 653 proteins [19]. A total of 814 proteins were identified from the articular cartilage tissue from normal donors and patients with osteoarthritis through SDS-PAGE followed by in-gel digestion [20]. Here, a total of 1510 proteins were identified, indicating the suitability of phenol extraction for clinical cartilage tissues.
In this study, 253 proteins were differentially changed between AVNFH and fracture patients. A total of 91 proteins were specifically detected in either the AVNFH patients or fracture patients (Supplementary Tables 2,3), further indicating the protein profile differences between them. From these 91 proteins, 81 proteins including caveolae-associated protein 3, tubulin polymerization-promoting protein 3, and Rho GTPase-activating protein 1, procollagen-lysine 2-oxoglutarate 5-dioxygenase 2, were specifically detected in the AVNFH patients; 10 proteins including hormone-sensitive lipase, perilipin-2, and metalloproteinase inhibitor 3 were reduced to undetectable levels in the AVNFH patients (Supplementary Table 2). Caveolae-associated protein is a principal component of caveolae membranes and functions as a scaffolding protein to organize and concentrate certain caveolin-interacting proteins within the caveolae membranes [21]. It is reported that caveolin-3 increases dramatically with age [22]. Tubulin polymerization promoting protein (TPPP) was identified as a disordered protein that affects microtubular system [23, 24]. It was reported that TPPP promotes the formation of synuclein filament, which is probably a crucial pathological process in certain neurological diseases [25]. The Rho GTPase-activating proteins are one of the major classes of regulators of Rho GTPases that are crucial in cell cytoskeletal organization, growth, differentiation, neuronal development and synaptic functions [26]. Procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD) genes are involved in fibrotic processes and tissue remodeling, and induced under hypoxic conditions [27]. These proteins were only detected in the AVNFH patients, indicating the abnormal expression of proteins associated with tissue or cytoskeletal remodeling, in the development of femoral head necrosis. Hormone-sensitive lipase is a key enzyme in the mobilization of fatty acids from acylglycerols in adipocytes as well as non-adipocytes [28]. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation [29]. The matrix metalloproteinases which degrade most of the components of the extracellular matrix, including fibrillar, non-fibrallar collagens, and fibronectin [30], and inhibitor of matrix metalloproteinases was drastically decreased in the AVNFH patients. These results indicate that accumulation of acylglycerols and degradation of extracellular matrix might happen alongside the development of AVNFH.
In this study, many ECM-receptor interaction or focal adhesion-related proteins including collagen alpha-1(VI) chain, thrombospondin-4, T-complex protein 1, chondroadherin, bone sialoprotein 2, and laminin subunit beta-2 were identified to be significantly changed between the AVNFH and fracture patients (Table 2). COL6A1 plays an important role in maintaining the integrity of various tissues, and alpha-1 subunit of type VI collagen is the encoded protein [31]. The thrombospondins are a family of extracellular calcium-binding proteins, of which, thrombospondin-4 binds specifically to collagenous extracellular matrix proteins via C-terminal domains [32]. T-complex protein 1 plays a role in the folding of actin and tubulin [33]. These proteins were significantly increased in AVNFH development, indicating the accumulation of extracellular matrix proteins and structural change of cartilage cells. Chondroadherin is a minor component of bone organic matrix and functions to promote attachment of osteoblastic cells to solid-state substrates [34]. Bone sialoprotein 2 functions to enhance fibroblast attachment in vitro [35]. Laminin subunit beta-2 is involved in sustaining the integrity of sarcolemmal basement membrane [36]. These proteins were significantly decreased in the AVNFH patients—an indication that, collapse of the microstructure is an inducer of AVNFH development. In future, the above identified key proteins which might be involved in AVNFH, need to be validated in a large sample size using other techniques such as western blot analysis and ELISA assays.
| ECM-receptor interaction | |||||
| Protein ID | Abbreviation | Description | Peptides | Ratio | p-value |
| O15335 | CHAD | Chondroadherin | 9 | 0.183091241 | 0.0005828 |
| P02458 | COL2A1 | Collagen alpha-1(II) chain | 33 | 0.381662001 | 0.0025274 |
| P11047 | LAMC1 | aminin subunit gamma-1 | 27 | 0.298013084 | 0.4224665 |
| P12109 | COL6A1 | Collagen alpha-1(VI) chain | 19 | 2.746900978 | 0.0004817 |
| P12110 | COL6A2 | Collagen alpha-2(VI) chain | 18 | 1.757307133 | 0.1391233 |
| P12111 | COL6A3 | Collagen alpha-3(VI) chain | 88 | 3.11349462 | 0.0000000628 |
| P21815 | IBSP | Bone sialoprotein 2 | 2 | 0.453476403 | 0.0120048 |
| P35443 | THBS4 | Thrombospondin-4 | 14 | 3.035757644 | 0.0029566 |
| P17987 | COMP | T-complex protein 1 subunit alpha | 6 | 1.645976288 | 0.0437059 |
| P55268 | LAMB2 | Laminin subunit beta-2 | 13 | 0.32725274 | 0.0173353 |
| Focal adhesion | |||||
| O15335 | CHAD | Chondroadherin | 9 | 0.183091241 | 0.0005828 |
| O43707 | ACTN4 | Alpha-actinin-4 | 17 | 0.532413165 | 0.0027161 |
| O75369 | FLNB | Filamin-B | 49 | 0.139170798 | 0.0006382 |
| P02458 | COL2A1 | Collagen alpha-1(II) chain | 33 | 0.381662001 | 0.0025274 |
| P11047 | LAMC1 | aminin subunit gamma-1 | 27 | 0.298013084 | 0.4224665 |
| P12109 | COL6A1 | Collagen alpha-1(VI) chain | 19 | 2.746900978 | 0.0004817 |
| P12110 | COL6A2 | Collagen alpha-2(VI) chain | 18 | 1.757307133 | 0.1391233 |
| P12111 | COL6A3 | Collagen alpha-3(VI) chain | 88 | 3.11349462 | 0.0000000628 |
| P12814 | ACTN1 | Alpha-actinin-1 | 17 | 0.547312925 | 0.0010154 |
| P21333 | FLNA | Filamin-A | 58 | 0.467185659 | 0.0038697 |
| P21815 | IBSP | Bone sialoprotein 2 | 2 | 0.453476403 | 0.0120048 |
| P35443 | THBS4 | Thrombospondin-4 | 14 | 3.035757644 | 0.0029566 |
| P17987 | COMP | T-complex protein 1 subunit alpha | 6 | 1.645976288 | 0.0437059 |
| P55268 | LAMB2 | Laminin subunit beta-2 | 13 | 0.32725274 | 0.0173353 |
In summary, a total of 1512 proteins were identified from cartilage tissues of the patients. Relative to the fracture patients, 255 significantly changed proteins were identified in cartilage tissues of patients with AVNFH. The results of functional categorization revealed that the significantly changed proteins were mainly involved in ECM-receptor interaction, focal adhesion, and glycolysis pathways. Of note, adipocyte enhancer-binding protein 1, cytoskeleton-associated protein 4, and ASPN protein were greatly decreased, however, antileukoproteinase, erythrocyte membrane protein, and lysozyme c were highly increased in patients with AVNFH.
The changed protein expression in the disease state is associated with many aspects of the pathogenesis of femoral head necrosis, such as increased proteolysis, lipid metabolism, immune response, degradation of extracellular matrix, and collapse of the microstructure. To our knowledge, this is the first time that a large portion of these proteins and their expression patterns have been identified in cartilage of femoral head necrosis patients. The findings of this study will help us to better understand the pathogenesis of AVNFH at the protein level and provide novel pathologic mediators and biomarkers for femoral head necrosis. This study also provides at least a proof-of-concept of the applicability of label-free proteomics in unraveling the mechanisms that underlie various diseases. In future studies, we would increase the sample size of the participants, concentrate on selected proteins and their underlying mechanisms as well as validate and quantify their levels by other known methods such as using ELISA kits and western blot analysis.
JH, FH, RNA, and XY made the major contributions to this study in the conception, design, drafting part of manuscript, and final revision. All authors read and approved the final manuscript.
The review board of the First Affiliated Hospital of USTC, University of Science and Technology of China approved this study (2020L L106) and participants provided signed consent forms.
We appreciate the help and support provided by the National Platform for Natural Medicine Active Components and the National Key Laboratory of Pharmacodynamics and the Platform of Chinese Medicine College.
This study was supported in part by Chinese Medicine Research Project (2016zy30) from Anhui Provincial Health and Family Planning Commission, the high-level personnel start-up fund from China Pharmaceutical University (No. 3154070004).
The authors declare no conflict of interest.
