Academic Editor: Graham Pawelec
Background: Metastatic melanoma (MM) represents a common malignancy with poor prognosis. Immune checkpoint inhibition (ICI), including PD-1 blockade, has been emerging as the popular therapeutic in MM for its durable treatment effect, but its response rate is still limiting. Methods: We comprehensively analyzed the associations between KMT2C somatic mutation and the tumor microenvironment as well as the ICI response of MM patients based on three published cohorts. Gene differential expression analysis between tumor samples with mutated and wild-type KMT2C was performed by DESeq2 package. Functional enrichment analysis was conducted by using clusterProfiler package. Kaplan-Meier was used to perform overall survival probability estimate through survival package and rms package was applied for the construction of nomogram model. Results: We report here that KMT2C is a potential biomarker for anti-PD-1 treatment in MM. This biomarker can be used for comprehensively analyzing its association with patients’ prognosis, tumor microenvironment and genomic features. Mutations of KMT2C profoundly altered expression of immune- and DNA replication-related genes in MM tumors. MM patients harboring KMT2C mutations showed significantly better overall survival (OS) after treatment with PD-1 monoclonal antibody as compared to wild-type KMT2C. Although KMT2C mutation has no significant influence on immune cell infiltration into MM tumors, the tumor mutation load and neoantigen load are indeed elevated in KMT2C mutated MM samples. This might represent a possible pathway through which KMT2C regulates the response of MM patients to anti-PD-1 treatment. Finally, we constructed a nomogram model by combing the independent prognostic factors, including KMT2C mutation, which could effectively predict the 1-year survival probability of MM patients after anti-PD-1 treatment. Conclusions: In conclusion, we report the role of KMT2C in anti-PD-1 treatment response regulation in MM for the first time. This may consequently be helpful for KMT2C personalized application.
Metastatic melanoma is a common malignancy with increasing incidence worldwide. Although the 5-year survival probability of localized melanoma could achieve 99% after surgical resection of the malignant lesions, the prognosis of metastatic melanoma (MM) is still poor with a 5-year survival probability of only about 20% [1]. Many therapeutics have been proposed for MM, such as adjuvant radio-/chemo-therapy before or after surgery, however, the efficiency has proven to be limited [2, 3]. Immunotherapy has long been a therapy of choice for multiple advanced malignancies. This type of treatment works through modulating tumor intrinsic immunity against tumor cells, such as chimeric antigens receptor-T (CAR-T) cell treatment in lymphomas and leukemias. The CAR-T treatment has shown encouraging anti-tumor efficiency [4]. Immune checkpoint blockade (ICB), mainly including anti-PD-1, anti-PD-L1, and anti-CTLA-4, represents the most popular immunotherapy method in the past decade. Some small-molecular inhibitors have been approved for clinical use by US Food and Drug Administration (FDA). For example, pembrolizumab which targets PD-1 and ipilimumab which targets CTLA-4 [5, 6]. MM is one of the most sensitive tumors in which ICB treatment and durable efficiency have been obtained in multiple clinical cohorts [7, 8, 9]. However, the response of MM patients to ICB is variable and identification of sensitive biomarkers is still urgently needed for the rational therapeutic schedule.
KMT2C, which is also known as MLL3, is a histone methyltransferase which specifically catalyzes the histone H3 lysine K4 mono-methylation at enhancer regions [10, 11]. KMT2C has been extensively studied for its role in genome stability regulation partially through the modulation of DNA damage repair [12]. However, controversial conclusions were drawn for its biological functions under various conditions. Larsson et al. [13] illustrated the significant association between KMT2C expression, repression and enhanced colorectal cancer cell growth. While Dawkins et al. [14] has found that depletion of KMT2C could profoundly inhibit the proliferation of pancreatic ductal adenocarcinoma cell lines. Chiappetta et al. [15] even showed the opposite effect of KMT2C knockdown on the migration capacity comparing primary and metastatic osteosarcoma cell lines. These observations indicate that KMT2C might execute biological functions in a context-dependent manner. Clinically, its mutation or aberrant expression has been widely associated with the prognosis or treatment sensitivity in multiple cancers [16, 17, 18, 19]. Our previous study [20] demonstrated the profound influence of KMT2C towards the sensitivity of breast cancer on chemotherapy through the regulation of genome stability. Response to ICB treatment has been closely related to genome instability in multiple cancers, however, the association between KMT2C and ICB response was rarely reported.
We report here for the first time the association between KMT2C mutation and ICB treatment including anti-PD-1 in MM. This might serve as a potential biomarker for personalized therapeutic schedule of MM patients.
Three MM cohorts used in this study included 68 MM samples from the Cancer Genome Atlas SKCM cohort (TCGA-SKCM), 144 MM samples from the study of Liu et al. [21] (DFCI2019), and 110 MM samples from the study of Allen et al. [22] (DFCI2015). Genome-wide mutation profiles of all the three MM cohorts and gene expression profiles of the TCGA-SKCM and DFCI2019 cohorts were accessible and were used for the analysis of this study.
DESeq2 function package [23] of R programming software version4.0.2 was applied
to screen differential expression genes (DEGs) in KMT2C mutated (KMT2CMut) MM
tumor samples. These were compared to KMT2C wild-type (KMT2CWT) samples in the
TCGA-SKCM cohort. Absolute value of log2 (Fold Change) (log2FC)
Functional enrichment analysis of DEGs was conducted by clusterProfiler function
package [24] of R programming software with the significant threshold of
p-value
Overall survival (OS) probability of MM patients after anti-PD-1 treatment in
DFCI2019 cohort was estimated via Kaplan-Meier method by using the survival
function package
(https://CRAN.R-project.org/package=survival)
of R programming software. Score test was applied to determine the significance
of OS probability difference among all MM patient groups with the threshold of
p-value
Nomogram represents a useful means of the prediction of survival probability at specific time points. In this study, we constructed a nomogram model for predicting the 1-, 2-, and 3-year OS probability after anti-PD-1 treatment using the rms function package (https://CRAN.R-project.org/package=rms) of R programming software and included the independent factors in the multivariate cox regression analysis.
Comparisons of quantitative variables, including tumor mutation burden (TMB),
neoantigen load (NAL), between KMT2CMut and KMT2CWT MM samples were conducted
using two-sided student t-test with the threshold of p-value
KMT2C has been reported to frequently mutate in multiple cancers. In this study, consistent high mutation frequency of KMT2C was observed in the three MM patient cohorts (11 out of 68 MM patients, 16.2% in TCGA-SKCM cohort, 18 out of 144 MM patients, 12.5% in DFCI2019 cohort, 16 out of 110 MM patients, 14.5% in DFCI2015 cohort). In addition to that, based on the TCGA-SKCM cohort, lollipop plot illustrating the distribution of KMT2C mutation sites across its protein functional domains are presented in (Fig. 1A). Lollipop plots of DFCI2019 and DFCI2015 cohorts are provided in Supplementary Fig. 1. It was observed that significant enrichment of mutations in the first PHD domain of KMT2C was obtained, which is consistent with the previous report [16].
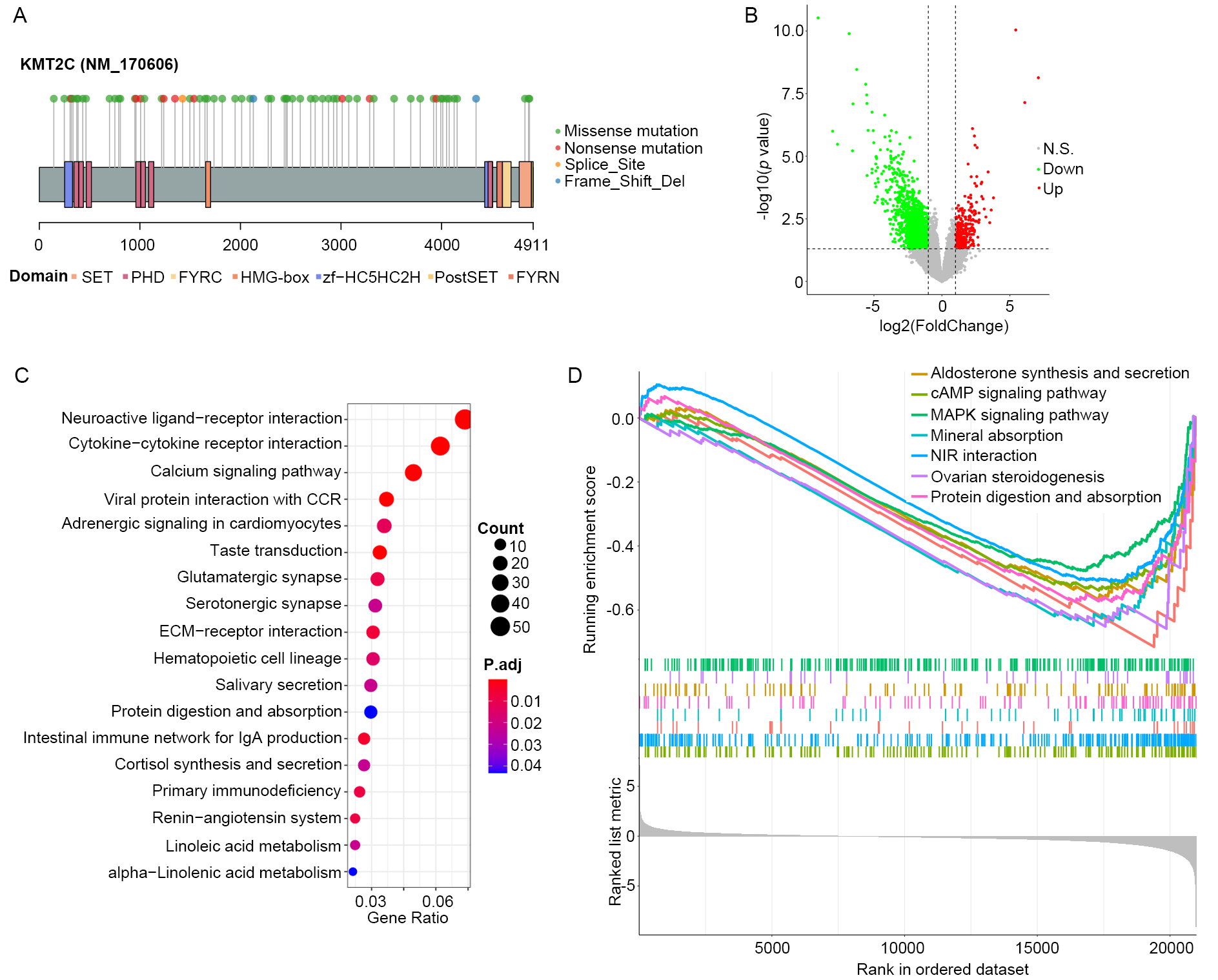 Fig. 1.
Fig. 1.KMT2C is functionally associated with immune regulation. (A)
Lollipop plot illustrating KMT2C mutations in MM samples in the TCGA-SKCM cohort
across function domains of KMT2C protein. (B) Volcano plot showing the result of
differential expression analysis between KMT2CMut and KMT2CWT MM samples in the
TCGA-SKCM cohort. Green and red dots represent significantly down- and
up-regulated genes in KMT2CMut compared with KMT2CWT MM samples, and grey dots
are nonsignificant genes. Vertical and horizontal dashed line indicate the
significant threshold of
To explore the functional consequence of KMT2C mutation, we screened the genes that were differentially expressed in KMT2CMut samples compared to KMT2CWT samples in the TCGA-SKCM cohort. The result showed that 1422 significantly down-regulated genes and 317 up-regulated genes were present (Fig. 1B). This is consistent with the transcriptional activation role of KMT2C as a histone methyltransferase. Functional enrichment analysis showed a total of 18 significantly enriched KEGG pathways (Fig. 1C) of the 1739 DEGs, including those that were closely related to tumor microenvironment status, such as cytokine-cytokine receptor interaction, ECM-receptor interaction, intestinal immune network for IgA production, and primary immunodeficiency among others. In addition to that, GSEA also identified seven significantly repressed KEGG pathways in KMT2CMut MM samples as shown in (Fig. 1D). These pathways were all cancer-related, e.g., cAMP signaling pathway, MAPK signaling pathway. These results reveal the potential role of KMT2C in cancer and immune regulation.
Investigation of the functional aspect of KMT2C mutations indicated its possible
influence on tumor microenvironment, which further prompted us to explore the
association between KMT2C mutation and immunotherapy response. Anti-PD-1
monoclonal antibody signifies one of the major immunotherapy methods which has
been extensively used in MM research. In this study, we investigated the
association between KMT2C mutations and prognosis of MM patients after anti-PD-1
treatment within DFCI2019 MM cohort. This included 144 MM samples that received
nivolumab or pembrolizumab, the two most common anti-PD-1 reagents. The result of
this study showed that KMT2C mutations are significantly associated with higher
OS probability after anti-PD-1 treatment when compared to KMT2C wild-type MM
samples (Fig. 2A). Some confounding factors might also be associated with the
prognosis of MM patients, such as gender, anti-CTLA4 treatment before anti-PD-1
treatment, metastatic sites, etc. To exclude the potential impact of these
confounding factors on the association between KMT2C mutation and response of MM
patients to anti-PD-1 treatment, we performed Fisher’s exact test. This test was
performed in order to establish if there was any significant difference in these
factors between KMT2CMut and KMT2CWT MM samples. As a result, only liver
metastasis showed significant difference between KMT2CMut and KMT2CWT MM samples.
In particular, KMT2CMut MM patients contained higher proportion of liver
metastasis samples than KMT2CWT MM patients (Fig. 2B). In addition to that, we
performed univariate cox regression analysis for all those confounding factors to
verify their associations with OS probability of anti-PD-1 treated MM patients.
Four factors in addition to KMT2C mutation, including brain metastasis, bone
metastasis, lactate dehydrogenase (LDH) level, and anti-CTLA4 treatment status
before anti-PD-1 treatment, were found to be potentially associated with OS
probability of anti-PD-1 treated MM patients (Fig. 2C). To determine if KMT2C is
an independent factor for the response to anti-PD-1 treatment of MM patients, we
performed multivariate Cox regression analysis by including all significant
factors in the univariate Cox regression analysis. As a result, KMT2C mutation,
brain metastasis, LDH level, and anti-CTLA4 status were statistically determined
as independent prognostic factors of anti-PD-1 treated MM patients with the
threshold of p-value
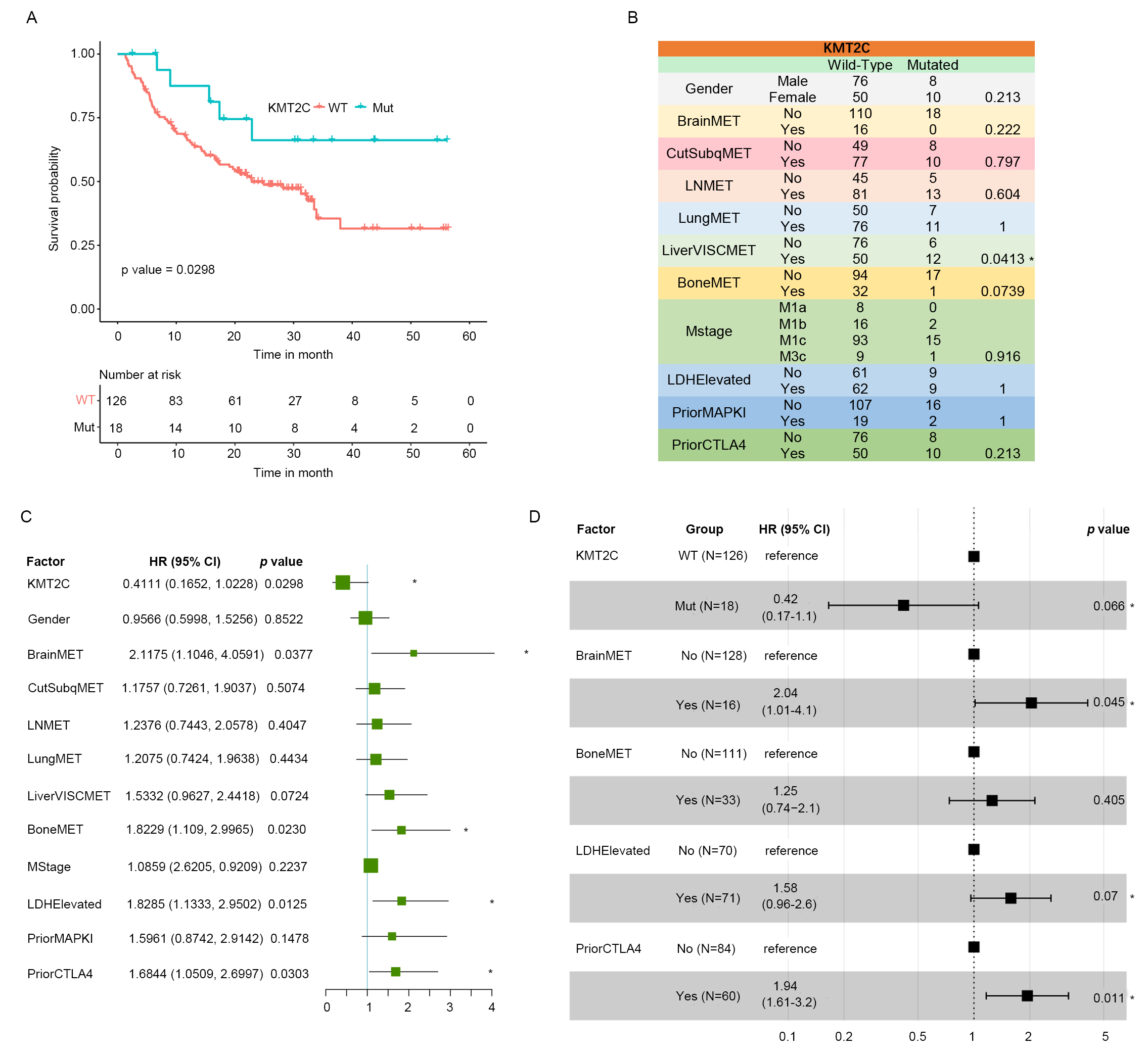 Fig. 2.
Fig. 2.KMT2C is an independent prognostic factor in anti-PD-1 treated
MM samples. (A) Kaplan-Meier plot of MM samples in the DFCI2019 cohort
stratified by the KMT2C mutate status. p-value was determined by score
test using the survival function package. (B) Confounding factor distribution
among KMT2CMut and KMT2CWT MM samples in the DFCI2019 cohort. p-value
was determined by Fisher’s exact test. BrainMET, Brain metastasis; CutSubqMET,
Subcutaneous metastasis; LNMET, Lymph node metastasis; LungMET, Lung metastasis;
LiverVISCMET, Liver metastasis; BoneMET, Bone metastasis; LDHElevated, LDH level;
PriorMAPKI, MAPK inhibition treatment status before anti-PD-1 treatment;
PriorCTLA4, anti-CTLA4 treatment status before anti-PD-1 treatment. (C) Forest
plot of the result of univariate cox regression analysis for the association
between KMT2C as well as other confounding factors and OS probability of MM
patients after anti-PD-1 treatment in DFCI2019 cohort. * indicates significant
association at the threshold of p-value
Tumor immune cell infiltration has been widely studied for its association with host intrinsic immunity against cancer cells as well as with cancer immunotherapy response. We hypothesized that regulation of immune cell infiltration into tumor mass might be a potential route through which KMT2C mutation influences the response of MM patients to anti-PD-1 treatment. To test this premise, we obtained the infiltration ratio of 22 immune cells of MM patients in TCGA-SKCM and DFCI2019 cohorts from the study of Charoentong et al. [25] and Liu et al. [21], respectively. The infiltration levels of the 22 immune cells in the tumor samples of MM patients are shown in the form of heat map in Fig. 3A,B. Macrophage M2, an immune-suppressed cell, showed significantly higher infiltration level than other cell types in MM tumors in both TCGA-SKCM and DFCI2019 MM patient cohorts. Immune activated cell macrophage M1 had relatively low infiltration level. Moreover, cytotoxic T cells, i.e., CD8 T cells, showed a modest infiltration level, which indicated that MM patients might intrinsically function against cancer cells if provided an appropriate tumor microenvironment. It was also observed that none of the 22 immune cells showed significant infiltration difference between KMT2CMut and KMT2CWT MM samples. In addition to that, we further tested if there were any significant differences in common immune checkpoint gene expression, including PD-1, PD-L1 and CTLA4, between KMT2CMut and KMT2CWT MM samples in the TCGA-SKCM and DCFI2019 cohorts. As a result, no gene except PD-L1 in the DFCI2019 cohort (KMT2CWT versus KMT2CMut: 2.50 versus 3.38, p-value = 0.029) showed significant difference between KMT2CMut and KMT2CWT MM samples in both cohorts (Fig. 3C,D). Those results indicate that KMT2C might influence anti-PD-1 treatment response through other pathways but not immune cell infiltration and checkpoint gene expression.
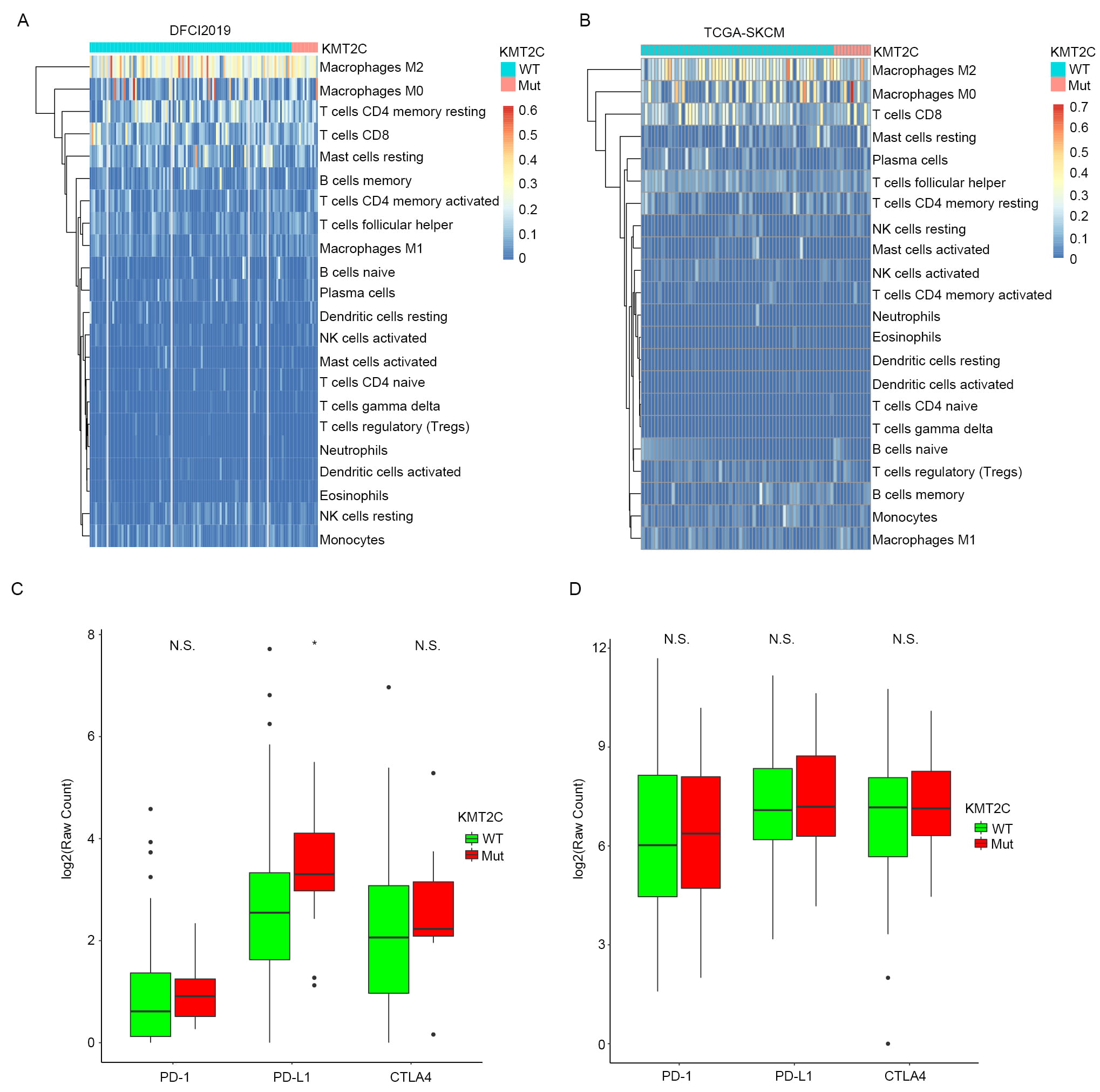 Fig. 3.
Fig. 3.Association between KMT2C mutation and immune cell infiltration
into MM tumor tissue. (A) Heatmap illustrating infiltration level of the 22
immune cells into tumor tissues of MM patients in DFCI2019 cohort stratified by
KMT2C mutate status. (B) The same as (A) but in the TCGA-SKCM cohort. (C) Boxplot
of the relative mRNA level of three immune checkpoint genes, including PD-1,
PD-L1 and CTLA4, in KMT2CMut and KMT2CWT MM samples in DFCI2019 cohort. N.S.
indicates not significant and * indicates significant difference at the threshold
of p-value
Tumor mutation burden (TMB) represents a widely used prognosis marker in many cancers and has been clinically used for estimating the response of targeted therapy or immunotherapy. Several factors have been closely linked to the TMB level through different pathways, including perturbation in the DNA replication process, DNA damage repair, etc. It has been previously reported that KMT2C is involved in regulating DNA replication as well as DNA damage repair pathways [26], such as homologous recombination. Based on these observations we investigated the association between KMT2C mutation and TMB in MM patients. TMB in this study was defined as the total number of nonsense and missense mutations per Mb genome. The result indicated that KMT2CMut MM samples show significantly higher TMB compared to KMT2CWT MM samples in all the three MM patient cohorts (Fig. 4A). Neoantigen is a type of antigen presented in the tumor cell surface and is highly individual-specific. Presence of neoantigen effectively increases the identification probability of tumor cells by cytotoxicity T cell and is closely associated with a better prognosis of multiple cancers [27, 28, 29]. In addition, the occurrence of mutation also endorsed that it contributes to the formation of neoantigen. TMB has been reported to be positively correlated with neoantigen load (NAL) in lots of studies, including our current study (Supplementary Fig. 2). Given these aspects, we further compared the NAL in the three MM patient cohorts between KMT2CMut and KMT2CWT samples. As expected, the NAL in KMT2CMut samples proved to be significantly higher than those KMT2CWT samples in all three cohorts (Fig. 4B). This indicates that the enhancement of NAL should be a potential path with which KMT2C mutation sensitizes the response of MM patients to anti-PD-1 treatment.
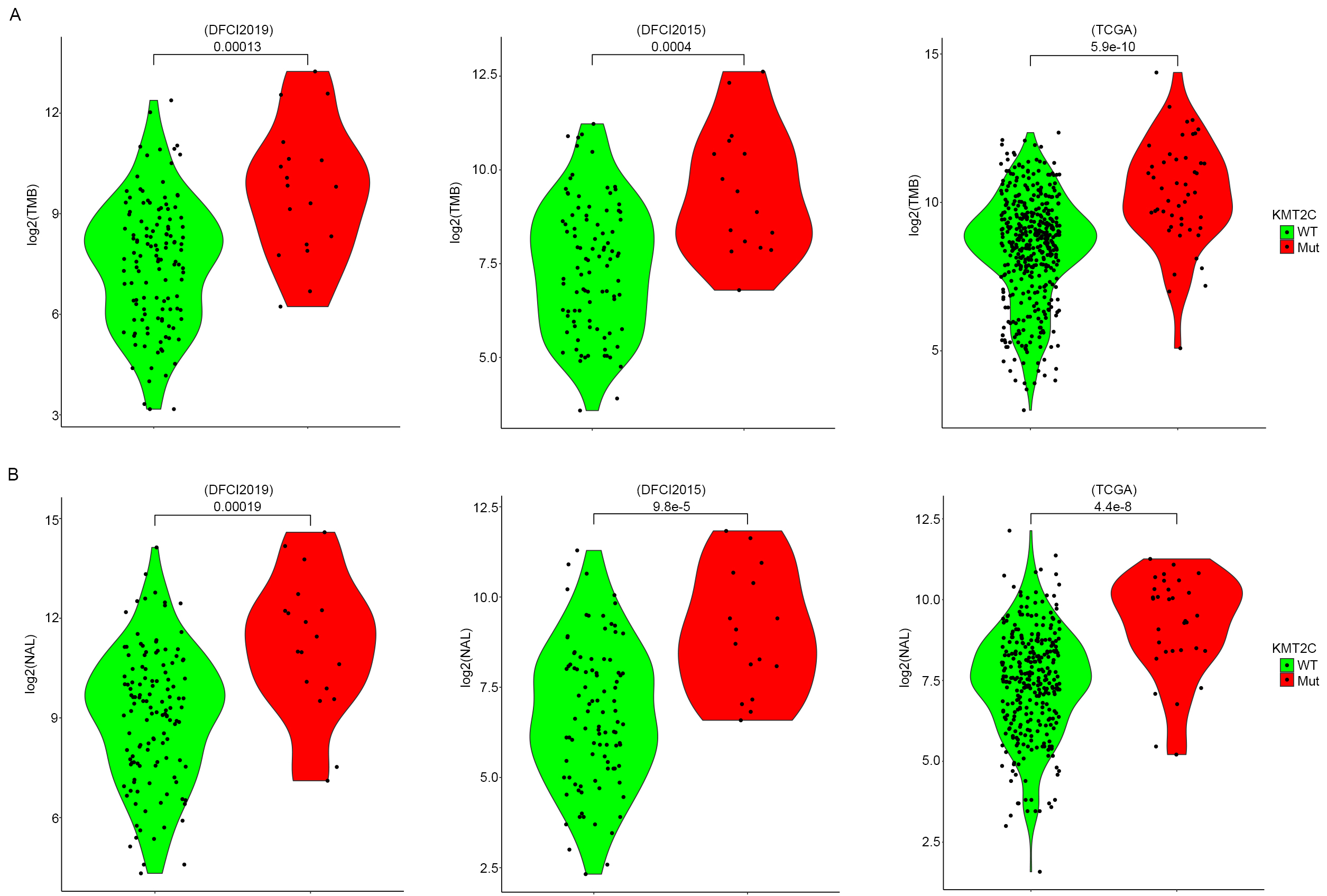 Fig. 4.
Fig. 4.Influence of KMT2C mutation on TMB and NAL of tumor tissue of MM patients. (A) TMB level (log2-based) of MM samples stratified by KMT2C mutate status in DFCI2019 (left), DFCI2015 (middle), and TCGA-SKCM (right) cohort. The p-value was determined by two-sided student t-test. (B) NAL level (log2-based) of MM samples stratified by KMT2C mutate status in DFCI2019 (left), DFCI2015 (middle), and TCGA-SKCM (right) cohort. The p-value was determined by two-sided student t-test.
Nomogram is widely used for predicting disease prognosis that consists of multiple relevant factors. In this study, we constructed a nomogram that predicted the 1-, 2-, and 3-year OS probability of MM patients after anti-PD-1 treatment in DFCI2019 cohort. This included the independent prognostic factors in the multivariate cox regression analysis, i.e., KMT2C mutation, brain metastasis, LDH level, and anti-CTLA4 status (Fig. 5A). It was found that KMT2C mutation, brain metastasis-free, low LDH level, and naïve anti-CTLA4 before anti-PD-1 treatment, were validated to be associated with higher 1-, 2-, and 3-year OS probability of MM patients after anti-PD-1 treatment. To evaluate the performance of the combined nomogram in predicting the OS probability of anti-PD-1 treated MM patients at different time points, the calibration curves for 1-, 2-, and 3-year OS probability were plotted to estimate the deviation between the actual and nomogram predicted OS probability. As a result, although the deviation was relatively large between the actual and nomogram predicted anti-PD-1 treated MM patient OS probability at 2- and 3-year, it was indeed very small at 1-year (Fig. 5B). Based on this study the nomogram might be useful in predicting the short-term OS probability of MM patients after anti-PD-1 treatment.
 Fig. 5.
Fig. 5.Nomogram for predicting the prognosis of anti-PD-1 treated MM patients. (A) Nomogram for the 1-, 2-, and 3-year OS probability prediction of MM patients after anti-PD-1 treatment in the DFCI2019 cohort by including the significant factors in multivariate cox regression analysis. (B) Calibration curves for estimating the performance of the nomogram model in predictin
Immunotherapy in the last decade has been revolutionarily developed and applied in multiple cancers [30, 31, 32]. MM is one of the most common scenarios that immunotherapy, particularly ICB is used for its intrinsic high TMB which underlies the highly activated tumor immunity [33, 34, 35]. Durable effects of ICB in MM have been widely obtained, but the low response rate largely impedes its extensive application. Identifying response biomarkers for ICB is currently in urgent need to accelerate the robust and rapid development of ICB application in cancer [36]. Here, we report KMT2C as a potential anti-PD-1 treatment marker in MM patients.
KMT2C is a well-known epigenetic regulator that plays important role in transcriptional regulation, specifically activation, by loosening chromatin structure through its catalytic role in mono-methylation of histone H3 lysine K4 (H3K4) [37]. Epigenetic regulation plays fundamental roles in lots of biological and clinical aspects [38, 39]. KMT2C was previously shown to contribute to the genomic stability and its mutation leads to the obvious TMB elevation in multiple cancers [20, 40]. In this study, a consistent positive association between KMT2C mutation and higher TMB in MM patients was observed, which also might result in the increase of NAL level. Neoantigen, which is produced by the mutation in exon part of the gene, is unlike the common antigen, such as carcinoembryonic antigen, which is highly specific to each individual and widely used for the design of personalized cancer vaccine [41, 42, 43]. In addition, high NAL would profoundly enhance the identification of cancer cells by cytotoxicity T cells and induce immune response for cancer cell removal, which is closely associated with better clinical manifestation, such as slow cancer progression and prolonged overall survival [27, 44]. Here we propose that KMT2C mutation contributes to a better anti-PD-1 response of MM patients which might be tightly related to its correlation with the elevation of TMB and later the NAL level. The role of KMT2C in immunotherapy response in other cancers was sporadically reported [45, 46], but the understanding of underlying mechanisms is very poorly understood and further studies are still needed.
In conclusion, we report here the potential role of KMT2C in the regulation of response of MM patients to anti-PD-1 treatment for the first time. This should be helpful for future studies regarding the sensitivity and extent of the clinical use of ICB treatment.
KX analyzed the data and wrote the manuscript. YP analyzed the data. WZ and XL proposed the study and revised the writing.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
