Academic Editor: Graham Pawelec
The title usage of Unde venisti ‘from where have you come’ is from a now dead language (Latin) that foundationally influenced modern English (not the major influence, but an essential formative one). This is an apt analogy for how both the ancient eukaryotic and eumetazoan functions of PGRMC proteins (PGRMC1 and PGRMC2 in mammals) probably influence modern human biology: via a formative trajectory from an evolutionarily foundational fulcrum. There is an arguable probability, although not a certainty, that PGRMC-like proteins were involved in eukaryogenesis. If so, then the proto-eukaryotic ancestral protein is modelled as having initiated the oxygen-induced and CYP450 (Cytochrome P450)-mediated synthesis of sterols in the endoplasmic reticulum to regulate proto-mitochondrial activity and heme homeostasis, as well as having enabled sterol transport between endoplasmic reticulum (ER) and mitochondria membranes involving the actin cytoskeleton, transport of heme from mitochondria, and possibly the regulation/origins of mitosis/meiosis. Later, during animal evolution, the last eumetazoan common ancestor (LEUMCA) acquired PGRMC phosphorylated tyrosines coincidentally with the gastrulation organizer, Netrin/deleted in colorectal carcinoma (DCC) signaling, muscle fibers, synapsed neurons, and neural recovery via a sleep-like process. Modern PGRMC proteins regulate multiple functions, including CYP450-mediated steroidogenesis, membrane trafficking, heme homeostasis, glycolysis/Warburg effect, fatty acid metabolism, mitochondrial regulation, and genomic CpG epigenetic regulation of gene expression. The latter imposes the system of differentiation status-sensitive cell-type specific proteomic complements in multi-tissued descendants of the LEUMCA. This paper attempts to trace PGRMC functions through time, proposing that key functions were involved in early eukaryotes, and were later added upon in the LEUMCA. An accompanying paper considers the implications of this awareness for human health and disease.
Progesterone (4-Pregnene-3,20-dione, hereafter: P4) receptor membrane component
(PGRMC) 1 (PGRMC1) is the archetypal member of a divergent eukaryotic branch of
the cytochrome b
We can consider cell and organismal biology at the level of a complex biological program, analogous to a computer program. The many stranded and even root level functionality of PGRMC proteins seems to affect the programmed response at many levels, from cellular to organismal, which in part reflects the evolutionary history of functional acquisition by PGRMC proteins. This constellation is permeated with still intangible potentialities for pharmaceutical intervention in disease processes (such as fertility, metabolic control, cancer, or neurological disorders), and perhaps modulation to extend healthy life. A conceptual understanding of the phenomenological interrelationships between PGRMC functions may be necessary to begin to target specific modularities, while leaving other perhaps critical functions intact. In that respect, it is necessary to first identify all PGRMC functionalities and their interrelationships, before being able to functionally stratify and finally safely pharmacologically address individual functions. To the extent that this is not yet possible, this work is conceptualized as a signpost.
This review attempts to orient the direction of future research by considering the overarching biological landscape, with the conviction that all PGRMC biology is potentially disease-relevant. However, much of the matter covered will be foreign to the medical researcher. Fundamentally, this work pursues the metabolism aspect of PRMC1, including consideration of age-related diseases such as cancer and diabetes, with special focus on the related cell and metabolic changes associated with the neuropathology of Alzheimer’s disease (AD) and possibly aging itself. These will be addressed in the accompanying paper [2] after this present paper builds a conceptual platform.
All cytb5 domain proteins contain a small heme-binding protein fold. The classical mammalian cytb5 proteins, of which humans have separate mitochondrial and endoplasmic reticulum (ER) species, are best characterized for contributing electrons to cytochrome P450 (CYP450) reactions, particularly in the process of steroidogenesis, although that is not their sole function [3]. The MAPR class of non-classical cytb5 proteins were originally identified as having a variably long peptide insertion between helices 3 and 4 of the canonical cytb5 domain fold [1], which forms a loop on the protein surface [4, 5] that I have dubbed the MAPR interhelical insertion region (MIHIR) [6, 7].
It is possible, but not established, that a MAPR protein was present in the last eukaryotic common ancestor (LECA) [8]. This paper will build an argument below that the MIHIRs of possibly the LECA MAPR protein [8], and of the PGRMC protein in the last eumetazoan common ancestor (LEUMCA) [7], were critically important for the origin of eukaryotes and eumetazoans, respectively. Eumetazoans (‘true animals’) are the group of animals including cnidarians (sea anemones, corals, sea pens, jellyfish, box jellies and Hydrozoa, such as Hydra) and bilaterally symmetrical animals.
Because the proposed PGRMC critical functions underpin many foundational aspects of modern mammalian biology, they are proposed to play substantive roles in human disease. This work represents a summary of current knowledge as well as conjectured hypothetical interpretations/interpolations. The author has attempted to make the boundaries between review and conjecture clear. Due to the large scope of the material covered, not all original work in the field has been directly cited, for which the author apologizes to omitted authors. Rather, reviews are cited in many instances. Please see original cited works in those reviews for underlying data.
Animals possess three types of MAPR proteins: PGRMC, Neudesin (NENF) and Neuferricin (NEUFC) [9, 10, 11]. All three families were already represented in the common ancestor of opisthokonts, the eukaryotic group that contains fungi and animals [7]. Humans possess one NENF, one NEUFC and two PGRMC proteins (Fig. 1A, Ref. [6, 7]). PGRMC1 and PGRMC2 arose by presumed gene duplication of an original pgrmc gene prior to the evolution of jawed fish [6, 12]. This paper will not focus on NENF or NEUFC proteins (about which relatively little is known [7]). Rather, it deals primarily with evolutionary events that occurred before the origin of chordates, and therefore prior to the divergence of PGRMC1 and PGRMC2. Accordingly, the paper will refer to PGRMC proteins unless specifically meaning either of vertebrate PGRMC1 or PGRMC2. Unless otherwise stated, any PGRMC amino acid numbering refers to the cognate human PGRMC1 positions.
 Fig. 1.
Fig. 1.Human MAPR proteins. (A) Schematic depiction of the most commonly reported membrane topology of four human MAPR proteins. PGRMC, NEUFC and NENF diverged prior to the common ancestor of fungi and animals. PGRMC1 and PGRMC2 genes diverged early in chordate evolution. TM, transmembrane domain. For NEUFC and NENF secreted to the extracellular this is a presumed cleaved signal peptide. The position of cytoplasmic positive residues presumed to orient the signal peptide in the endoplasmic reticulum is indicated. Alternative topologies of the MAPR domain of each protein are known. For more detail see [6, 7]. (B) Comparison of human PGRMC1 and PGRMC2 (UniProt identifiers O00264 and O15173, respectively). PGRMC1 tyrosines 107, 113, 139 and 180, and regulatory phosphorylation sites T178 and S181 are shared with PGRMC2. SH2 and SH3 depict predicted target binding motifs for proteins containing SH2 domains and SH3 domains. The PGRMC1 62-66 SH3 motif is absent from PGRMC2. Binding sites for other proteins, as predicted by Scansite 4.0 (https://scansite4.mit.edu/), are indicated to demonstrate that although the proteins are similar, amino acid divergence can also be expected to lead to alternative biological functions. Other conventions follow (A).
It can be expected that the two structurally similar PGRMC1 and PGRMC2 proteins exhibit potential functional overlap, but also specific differences (Fig. 1B). Although an analysis of the relationship between PGRMC1 and PGRMC2 merits attention, I will not attempt to address that here. Rather, I will focus on the proposed role of an ancestral MAPR protein in eukaryogenesis, and much later of the proposed involvement of PGRMC in the origins of the gastrulation organizer, and the system of eumetazoan body pattern formation that it may have foundationally enabled. The accompanying paper [2] will consider how this combined biological panoply is relevant for our mechanistic understanding of several modern clinical pathologies.
PGRMC1 was cloned in a variety of different functional contexts in different organisms, including as part of a membrane P4 receptor protein complex, as a dioxin-inducible neural gene, as a protein involved in adrenal steroidogenesis, and as a ventral midline antigen involved in axon guidance during embryological genesis of the central nerve cord. It also emerged that the P4 receptor function of PGRMC1 was important in the female reproductive system, which included expression in both oocytes and their granulosa feeder cells (please refer to original literature cited in previous 2007 reviews [5, 13]).
Granulosa cells were found to exhibit a P4-dependent protection against apoptosis [14]. This P4-induced vitality-conferring function of PGRMC1 was later shown to protect against P4 withdrawal-induced cell death in both granulosa and luteal cells [15] and in MES-SA uterine sarcoma cell [16]. PGRMC1 was later shown to simultaneously decrease proliferation and increase resistance to doxorubicin-induced cell death in a P4-dependent manner in Ishikawa endometrial cancer cells [17], MDA-MB-231 breast cancer cells [18] and MIA PaCa-2 pancreatic cancer cells [19]. This affect appears to be mediated by PGRMC1-dependent CYP450 modification of doxorubicin [4].
Progesterone actions were historically first discovered to be mediated by the nuclear (i.e., classical) P4 receptor (PGR) through its isoforms (PGR-A and PGR-B). Note, for the perspective of this present paper, that conventional steroid receptors and their ligands (the steroid hormones which are synthesized from cholesterol) are a eumetazoan animal invention [6]. PGRMC1 became known as one of the non-classical P4 receptors, which include PGRMC1/2 as well as several G protein-coupled receptors [13, 20, 21, 22, 23, 24, 25]. Furthermore, nuclear PGR expression is reduced by PGRMC1 and PGRMC2 double knockout [26], so that nuclear PGR steroid responsiveness can be considered as evolutionarily and functionally subservient to that of PGRMC. For a recent review on the role of all steroid receptors in cancer, see Masi et al. [27].
Although PGRMC1 had been suggested to be identical to low affinity glucocorticoid binding site (LAGS), Marek et al. [28] demonstrated that this was not the case. Indeed, an inverse situation may apply, where more of the response to the progestin medroxyprogesterone acetate (MPA) is mediated by the glucocorticoid receptor than by PGRMC1 [29]. Glucocorticoid receptor may even drive the elevation of PGRMC1 expression in response to MPA [30].
Understanding PGRMC functions has clearly developed as an intriguing story, which is embellished by citation of some early works relevant to this paper, whose importance the author did not emphasize in his 2007 review. Firstly, PGRMC-like proteins were widely distributed among eukaryotes (but none were known from prokaryotes) [5]. Secondly, the yeast Saccharomyces cerevisiae PGRMC homologue Dap1 (‘damage response protein related to membrane-associated P4 receptors’) was involved not only in DNA damage repair, but also with the lanosterol-14-demethylase reaction, where, like classical cytb5 proteins, its heme iron was proposed to contribute to the CYP450 redox reaction [31]. In this respect, note that McGuire et al. [32] argue that PGRMC1 is unlikely to act as an electron carrier in catalyzed reactions due to its pentacoordinate heme chelation that involves a negatively charged tyrosinate (an ionized tyrosine hydroxyl group) interaction with the iron heme atom. It is not immediately clear that this deductive reasoning is sound. It is conceptually feasible that specialized enzyme active sites could well have evolved to facilitate redox reactions involving PGRMC heme iron electron carriers. However, the proposal of McGuire et al. [32] may indeed be correct.
Kabe et al. [4] later demonstrated that only heme bound but not the bacterially-expressed apo-PGRMC1 N-terminally-truncated MAPR domain (containing a Y113F mutation which was thought to ameliorate heme interaction) interacts with CYP51A1. Recently, McGuire et al. [32] showed that a Y113F mutant of full length PGRMC1 binds heme similarly to the wild-type PGRMC1 sequence, whereas a Y113F, K163A, Y164F (3X MUT) PGRMC1 did not bind heme. Wild-type PGRMC1 as well as the Y113F and 3X MUT PGRMC1 all bound to and stabilized the protein levels of diverse cyP450s, which led to elevated cyP450 activity. Therefore, PGRMC1 binding to cyP450 is independent of the ability to bind heme. Since the cyP450 interaction of PGRMC1 with CYP51A1 is the oldest known conserved function, possibly related to eukaryogenesis, it is tempting to speculate that this interaction involves the contiguous stretch of surface residues on the MAPR domain (that are scattered throughout the MAPR domain primary sequence but form a single surface feature) which shares strongest conservation with prokaryotic cytb5M (which we named as MAPR-like cytb5) proteins from which MAPR proteins originated [8, 33].
Lanosterol-14-demethylase (Erg11p in S. cerevisiae and Schizosaccharomyces pombe, CYP51A1 in mammals) is the cyP450 which catalyzes the very first modification of lanosterol. The latter is the first sterol produced by squalene cyclase from the mevalonate pathway (Fig. 2, Ref. [8, 34, 35]; Fig. 3, Ref. [36, 37, 38]) [36]. Lanosterol demethylase is the most conserved cytochrome P450 in eukaryotes, catalyzing the 14-demethylation of sterols in plants, fungi, and animals [39, 40]. Espenshade’s group showed that the Dap1 (PGRMC)/lanosterol-14-demethylase interaction also operated in the fission yeast S. pombe [37]. Much later, we showed that both yeast Dap1 proteins are PGRMC-like proteins, rather than NENF-like or NEUFC-like. Both fungi possessed just a single PGRMC-like MAPR gene, with no known NEUFC or NENF counterparts, although the ancestral opisthokont common ancestor of fungi and animals possessed all three gene families [7].
 Fig. 2.
Fig. 2.Early steroid biosynthetic reactions require molecular oxygen. (A) Squalene (an isoprenoid product of the mevalonate pathway) conversion to lanosterol occurs in two reactions. In the first reaction squalene epoxidase (also called squalene monooxygenase) produces a 2,3 epoxide in a reaction that requires molecular oxyen. In the second reaction oxidosqualene cyclase (OSC), also called lanosterol synthase, produces lanosterol (the first sterol). OSC is related to bacterial squalene-hopene cyclases (SHCs). Unlike SHCs, which can react with a variety of substrates, OSCs exhibit strong substrate selectivity for 2,3-epoxides such as oxidosqualene [34, 35]. Lanosterol synthesis is proposed to have arisen from a modified hopanoid synthetic pathway in response to the presence of oxygen following the appearance of photosynthesis. (B) Lanosterol is C14-demethylated by CYP51A1 (also known as lanosterol-14-demethylase) and PGRMC1 (C14 is highlighted). The reaction is shown in the box (Source: UniProt Q16850), highlighting the requirement of oxygen as substrate. All enzymes involved in MVP to FF-MAS production are from bacterial rather than archaeal origin in eukaryogenesis [8].
 Fig. 3.
Fig. 3.Steroid biosynthesis in mammals. Cholesterol is produced from
lanosterol by two major pathways: the Bloch (black arrows) and the modified
Kandutsch-Russell (red arrows) pathways. The image shows the position of the
CYP51A1 reaction that involves PGRMC1 [37]. Squalene is produced by the
mevalonate pathway (MVP) and converted to lanosterol (Fig. 2A). CYP51A1 catalyzes
the first modification of lanosterol (Fig. 2B). Requirement for molecular O
Finally, in breast cancer cells Asperger et al. [41] have recently demonstrated that PGRMC1 interacts with multiple components of the pathway of metabolites from mevalonate to lanosterol, including Acyl-CoA desaturase, squalene synthase and squalene monooxygenase. They also confirmed the CYP51A1 interaction [41]. Those enzymes include all of those in Fig. 2 that convert the mevalonate pathway product of squalene into eukaryotic-specific steroids, and which were probably acquired by the LECA.
In summary, PGRMC involvement with the CYP51A1 reaction was ancestral to opisthokonts, and very likely also to eukaryotes (which has not been systematically examined). Involvement of a PGRMC-like protein in the regulation of the lanosterol-14-demethylase CYP450 reaction in the ancestor of yeasts [31, 37], and the regulation of the upstream steroidogenic SREBP/Insig-1/SCAP (sterol regulatory element binding protein/ insulin-induced gene 1 protein/SREBP cleavage activating protein) complex by PGRMC1 [42, 43] jointly suggest that the PGRMC sterol response should be far more ancient in eukaryotic biology than the steroid responses induced in animals by classical/nuclear steroid receptors. Note that the Insig/SCAP/SREBP complex is stabilized by 25-hydroxycholesterol binding to Insig [44], and 25-hydroxycholesterol also exhibits low affinity binding to the PGRMC1 interaction partner TMEM97 [45]. The role of PGRMC1 in the Insig/SCAP complex remains unexplained. McGuire et al. [32] have suggested that PGRMC1 could stabilize other proteins in addition to cyP450s. It is interesting to speculate that PGRMC1 could stabilize Insig, conceivably by preventing ubiquitination, in the presence of 25-hydroxycholesterol. It will be interesting to see whether TMEM97 is associated with the Insig/SCAP/PGRMC1 complex, and whether such complexes contain SREBP.
In clinical breast ductal carcinoma in situ, we found (1) that PGRMC1
phosphorylation status depended upon estrogen receptor (ESR
 Fig. 4.
Fig. 4.PGRMC1 inductions precedes GLUT1 in some DCIS hypoxic cells. Induction of PGRMC1 and GLUT1 in ductal carcinoma in situ (confocal microscopy, adapted from [46]). Some PGRMC1-positive cells have no GLUT1 (1). Other GLUT1-negative cells have no (2) or peri-nuclear (3) PGRMC1, whereas in regions of high GLUT1 expression many cells exhibit cytoplasmic or even possibly nuclear PGRMC1 (4), suggesting that nuclear PGRMC1 may be involved in the induction of the GLUT1 gene. This speculation requires further investigation.
After more than a decade of research hiatus in that area PGRMC1 has subsequently
been shown by us and others to indeed affect glycolysis [49, 50] and
gluconeogenesis [51], and to regulate glycogen synthase kinase-3
Hif-1 first appeared in the common ancestor of placozoans and eumetazoans [53]. G-protein-coupled receptors (GPCRs) were present in the LECA, but underwent a radiation with the origin of bilaterians [54]. The oxygen-dependent induction of steroidogenesis may represent an ancient and Hif-1-independent mechanism of metabolic regulation, as below elaborated. If true, any relationship of this ancient system to Warburg metabolism remains uncertain and requires investigation.
Other emergent functional contexts for PGRMC1 since 2007 include roles in cell
cycle regulation, particularly in controlling the G
 Fig. 5.
Fig. 5.PGRMC1 as a potential regulatory nexus hub protein. The figure depicts roles for PGRMC1 at the center of a signal network involving energy metabolism (glucose, amino acids [tryptophan is pictured] and fatty acids), P4 response, lipid regulation, heme and CyP450 biology, regulation of metalloproteases, membrane trafficking, cell cycle and mitotic spindle, extra-cellularization by the exosome microvesicle pathway, induction of angiogenesis via VEGF, gene regulation in the nucleus, sigma-2-receptor activity, as well as functions such as autophagy, etc. as listed in the boxes. For details of biological associations refer to the discussion in the main text and [47]. Modified from Fig. 4 of [47], with permission from Elsevier, BBA. Reviews on cancer. http://www.sciencedirect.com/science/journal/0304419X (License Number 4938620747881). Generation of the PGRMC1 4X8Y [4] dimer image has been described [66].
This present work is largely inspired by the interpretative implications of
several recent results from the author’s collaborative research results. We found
that altering PGRMC1 phosphorylation status changed the abundance of a suite of
proteins involved in energy metabolism, of which many were mitochondrial. This
was associated with changes in mitochondrial form and function (glycolysis,
O
Espenshade’s group had previously demonstrated that the S. pombe pathway acted as an oxygen sensor, in a mechanism involving sterol levels [68]. The concept of PGRMC-like proteins being related to oxygen levels will play a central role in the hypothesis developed here, so please note this seminal finding. This seemingly essential eukaryotic function (proposed to have been essential for early eukaryotes because of wide phylogenetic distribution and conserved function) suggests that the protein had played an important role in the evolution of early eukaryotes: if not in the LECA, then at least sometime prior to the last Opisthokont common ancestor.
The answer to the whether MAPR were involved in eukaryogenesis remains: “maybe”. Because of the small size of the protein and the large evolutionary distances involved, the answer could not be concluded with confidence. Either MAPR/PGRMC-like proteins with eukaryotic-like tyrosinate heme chelation were acquired by a group of bacteria by horizontal gene transfer from some eukaryote, or a member of that same class of bacterial proteins probably contributed to eukaryogenesis [8]. This paper argues according to the latter scenario. However, note that this assumption is not unambiguously supported by the available data because of the small size of the MAPR domain and the large evolutionary distances involved [8].
The next issue concerns the much later evolution of the animal gastrulation organizer. PGRMC1 contains two putative tyrosine phosphate acceptors that are thought to form the centers of Src homology 2 (SH2) target motifs (not SH2 domains, but places where proteins with SH2-domains could bind if PGRMC1 were tyrosine phosphorylated) [5, 69]. There has been some literature confusion about the presence of PGRMC1 SH2 domains, which do not exist. Tyrosine kinases and SH2 domains that bind to phosphorylated tyrosines evolved in the unicellular Opisthokont lineage leading to animals [70, 71, 72, 73]. We found that the PGRMC1 tyrosines (Y139 and Y180; Fig. 1) were acquired in evolution by the LEUMCA [7] which is recognized as the first organism that can be reconstructed to have possessed a functional gastrulation organizer [74, 75, 76, 77]. This theme will be developed below after we have considered the properties of the gastrulation organizer in more detail.
Iron is an essential element for all life on earth due to its redox properties as well as its electropositive ability to polarize atoms during reaction catalysis. It is used in chelated ligand interactions, as sensors for various substances, and in enzyme catalysis [78]. Paradoxically, this essential element is also highly deleterious. In free solution it can generate toxic reactive oxygen species via Fenton chemistry [79].
The most ubiquitous solution to this biological dilemma, found in all branches of life, is to chelate iron atoms into a porphyrin ring to form heme. Like the terrors of fire being tamed by a hearth, the advantageous properties of iron can thereby be biologically harnessed without suffering the ravages of Fenton chemistry. Heme manifests its influence on biological processes by acting as a prosthetic group for, e.g., proteins of the cytb5 domain family (including MAPR proteins like PGRMC1), other cytochromes (e.g., CYP450s) and their reductases, kinases, transcription activating factors, transcriptional repressors, ion channels, miRNA processing proteins, and globins, which together act as electron carriers for the enzymatic oxidation or reduction of substrates, mediate oxygen sensing, gas synthesis (e.g., CO), transcriptional regulation, iron homeostasis, oxidative stress response, mitochondrial biogenesis, circadian rhythms, or influence cell cycle progression and proliferation. The latter are of course directly critical to disease [78, 80, 81, 82].
Fungi, animals, and alphaproteobacteria (related to mitochondria: see below) synthesize heme via a C4 (“Shemin”) pathway, whereas archaea, all other bacteria (with few exceptions), and plants (which synthesize heme using a chloroplast pathway that arose via an independent bacterial endosymbiotic event) employ the evolutionarily older C5 (glutamate) pathway [80, 83]. Clearly, eukaryotic heme synthesis was inherited by opisthokonts (the group including fungi and animals) from the proto-mitochondrial alphaproteobacterial endosymbiont during eukaryogenesis. Enzymes for the ancestral archaeal host cell Shemin heme synthetic pathway were lost during the eukaryogenic process.
Free porphyrin and heme are toxic, and so they exist as protein prosthetic groups whose abundances are carefully regulated [80]. Heme oxygenase-1, which initiates heme degradation, is anti-inflammatory and is induced under conditions of oxidative stress (For review: [84]). Importantly for our theme: (1) multiple proteins involved in mitochondrial citric acid cycle and oxidative phosphorylation electron transport chain have heme prosthetic groups [78, 80], making heme essential (and regulatable) for mitochondrial function, (2) mitochondrial PGRMC1 associates with and regulates ferrochelatase, the last and rate-limiting reaction of heme biosynthesis [32, 85], (3) the redox-sensitive change in affinity of PGRMC1-bound heme led to speculation that it may act as a heme chaperone from its site of synthesis in the mitochondrial outer membrane to other subcellular locations [67, 85]. Indeed, PGRMC2 has subsequently been demonstrated to exhibit precisely such a chaperone function to the nucleus, where it regulates genes related to mitochondrial metabolism [82].
When PGRMC1 was discovered to be a CYP450-interacting cytb5 domain protein, it
was categorized by annotation data bases as an ER protein and was probably not of
much interest to pharmaceutical companies in the early 2000s. However, PGRMC
proteins are not your average cytb5 proteins. Tyrosinate heme chelation is unique
to the MAPR family and a few bacterial sequences (to be discussed later) among
cytb5 proteins [8]. It leads to a characteristically lower redox potential of the
heme, and lower affinity for ferrous/reduced heme in the yeast PGRMC-related Dap1
protein [86], in PGRMC1 (K
 Fig. 6.
Fig. 6.Possible MAPR origins and eukaryogenesis. (A) The lower
affinity of tyrosinate heme chelating MAPR proteins for ferrous/reduced heme [86]
imply that at least two functional states exist, each of which may hypothetically
exert discrete suites of functions. Site of PGRMC1 interactions with heme from
the 4X8Y crystal structure [4] are indicated by arrows and the color key. (B) A
schematic diagram of human PGRMC1 showing the positions of helices (H) and
The ‘one-hit chemistry’ that can be deductively associated with reduction and disassociation of heme after a PGRMC/CYP450-mediated reaction should lead to the presence of separate heme-bound PGRMC and heme-free apo-PGRMC, which could have dramatically different suites of functions (Fig. 6A), and which has received scant attention. The jettisoned heme must also be transferred to another as-yet unknown protein, which will change its properties. Note once more the argument of McGuire et al. [32] that tyrosinate-chelated heme iron is unlikely to act as an electron carrier.
The level of in vivo PGRMC1 heme occupancy was determined to approximate 0.6 [89], suggesting that populations of holo-PGRMC1 (with heme) and apo-PGRMC1 (without heme) exist. Lee et al. [51] recently argued for a discrete function of “PGRMC1 monomer” involved in glucose regulation after treatment with the small molecule AG-205, which should displace heme. McGuire et al. [32] also identified PGRMC1:cyP450 interactions and cyP450 stabilizations that were independent of heme binding. As will be developed below, the existence of heme-dependent and -independent functions may lie at the crux of both PGRMC1 and eukaryotic biology, with momentous implications for human disease. Lee et al. [51] do not define what is meant by “PGRMC1 monomers”. Nor do they acknowledge that AG-205 effects may be PGRMC1-independent, as explored in the next section.
Please note for the remainder of this manuscript that AG-205 has traditionally been regarded as a PGRMC1-specific inhibitor. For historical background on AG-205 see [90]. We have previously noted the absence of evidence that AG-205 was indeed specific for PGRMC1 [47, 91]. It has since been shown to cause the formation of large cytoplasmic vesicles in cells that have a double knockout of the pgrmc1 and pgrmc2 genes, and therefore does not exclusively affect PGRMC1 [90]. Kabe et al. [92] could not physically detect binding of AG-205 to either apo- or heme-bound PGRMC1. Furthermore, Thieffry et al. [93] newly report that AG-205 causes the induction of genes involved in sterol synthesis, including that for INSIG1 protein which forms a complex with PGRMC1. They use small interfering RNA (siRNA) attenuation of each of all four human MAPR proteins (PGRMC1, PGRMC2, NENF and NEUFC) to show that AG-205 induction of INSIG1 occurred after attenuation of each MAPR protein alone, or in combination, leading to the conclusion that AG-205 does not specifically target PGRMC1 or any human MAPR protein.
While it is possible that Thieffry et al. [93] are correct, and their study is welcomed and to be applauded, there remain several caveats. Their reasoning is based upon the assumption that PGRMC1 represents a single function, whereby removing PGRMC1 protein would have the sole effect of removing that single function. If the scenario of Fig. 6A is true, where heme-bound and apo-PGRMC1 could have alternative functions, perhaps only one of which could be modulated by AG-205, then removing PGRMC1 protein would not be expected to display identical phenotype to inhibition by AG-205. For instance, if the “Functional suite 2” of Fig. 6A were activated by AG-205, then PGRMC1 knockdown would not be expected to mimic AG-205 treatment because all PGRMC1 functions would be absent. Even if residual low levels of PGRMC1 remained, the AG-205-dependent effect would still require AG-205 to activate that function (which is possibly what Thieffry et al. [93] observed). Similar caveats apply to any alternative functions that could be regulated by PGRMC post-translational modifications.
Additionally, none of the siRNA-mediated MAPR attenuations achieved by Thieffry et al. [93] were absolute. It remains possible that low levels of residual PGRMC1 could still have mediated the observed AG-205-dependent effects through high affinity interaction partners which were able to recruit the low levels of PGRMC1 proteins present. Given the relatively close relationship between PGRMC1 and PGRMC2, and the proposed ancient nature of the eukaryotic PGRMC-dependent steroidogenic regulation in question, this is important for the single PGRMC1 knockdowns, where PGRMC2 could conceivable have fully compensated. Problematically, the absolute levels of PGRMC1 and PGRMC2 downregulation were less pronounced when all four anti-MAPR siRNAs were introduced simultaneously. It is conceivable that residual levels of PGRMC1 and/or PGRMC2 could still have led to the AG-205-dependent increase of INSIG1 levels (See Fig. 8 of Thieffry et al. [93]). Repeating the study design of Thieffry et al. [93] using MAPR gene knockouts should provide definitive evidence. In the meanwhile, it is prudent to acknowledge the advice that AG-205 should not be considered a specific PGRMC1 antagonist [47, 90, 91, 93], and to carefully reconsider any published conclusions which have relied upon this assumption.
Thieffry et al. [93] also propose that the steroidogenic biology attributed to PGRMC1 is largely caused by the ability of AG-205 to induce steroidogenic genes. This is unlikely to be true. PGRMC1 binding to the INSIG1/SCAP complex [42] is independent of AG-205, as is the regulation of CYP450A1 [31, 37] (Fig. 2). Based upon such reasoning the initial proposal that PGRMC1 was involved in steroidogenesis was made before the discovery of AG-205 [5, 46].
Furthermore, in female pgrmc1 heterozygous knockout (hetero KO) mice, which had correspondingly low PGRMC1 levels, estrogen levels were lower than in control mice [94]. Most estrogen synthesis occurs in the ovaries, with some non-ovarian synthesis. When hetero KO mice were ovariectomized then estrogen levels were higher than in control mice, implying that low PGRMC1 levels promote higher estrogen synthesis in extra-ovarian tissues [94]. These results associate PGRMC1 with steroidogenesis independent of the activity of AG-205. If we accept that PGRMC1 does affect steroidogenesis, it becomes extremely interesting to consider why AG-205 might be able to regulate the same pathway(s) in the absence of PGRMC1.
In another recent publication that assumes AG-205 is specific for PGRMC1, Sun et al. [95] observed that AG-205 prevented neuronal resistance to hypoxic ischemia by preventing activation of NF-kB signaling and the BDNF/PI3K/AKT pathway. They concluded that PGRMC1 exerts a neuroprotective role following brain injury. The proposed mechanism associating PGRMC1 with both neuronal and hypoxic biology would align excellently with the main hypothesis of this present paper and may also be correct. Indeed, PGRMC1 was previously known to be part of an induced neuronal regenerative tissue repair and remodeling response following brain and spinal cord injuries in rats [96, 97, 98], and PGRMC1 changes phosphorylation status as part of the response to chemical ischemia of the brain [99]. However, there was total disregard by Sun et al. for the possibility that AG-205-mediated effects could operate independently of PGRMC1. As such, none of the published evidence demonstrates the claim of their title that inhibition of PGRMC1 aggravates cerebral ischemic damage caused by hypoxia [95]. Journal authors, reviewers and editors must clearly be more critical when assessing conclusions based upon the use of AG-205.
Kabe et al. [4] have suggested that PGRMC1 forms a heme-mediated dimer. They presented convincing biophysical evidence of dimeric properties of bacterially expressed protein, and they solved the structure of a PGRMC1 dimer of bacterially expressed proteins, with contacts between heme rings of adjacent molecules. This led to the suggestion that PGRMC1 bound to ferrous heme could act as a sensor for carbon monoxide (CO), which cannot bind to ferric heme [4, 100]. They demonstrated the formation of ferric-heme-bound bacterially expressed PGRMC1 dimers in vitro, whereas dimerization was attenuated by addition of CO [4].
Some doubt remained (for this author at least) about the actual presence of a dimer in mammalian cells, because their bacterially-expressed protein contained an unfortunately designed N-terminal deletion of a few conserved positive residues from the PGRMC1 MAPR domain, and replaced these with four amino acids from the pGEX plasmid vector GST (glutathione-S-transferase)-fusion protein cloning linker region. Regrettably, some of the foreign N-terminal GST-linker residues make direct contact with the heme molecules in the crystal structure [4], which could artefactually stabilize a heme-dependent dimer for the bacterially expressed fusion protein. The bacterially expressed dimer was also stabilized by a cysteine bridge involving C129, which would not be favored in the reducing environment of mammalian cytoplasm.
The sole evidence offered by Kabe et al. [4] for the existence of a dimer in mammalian cells involved the ability of PGRMC1 to interact with the epidermal growth factor receptor (EGFR). Mutation of Y113, which is required for heme chelation, prevented interaction with EGFR by N-terminally truncated PGRMC1. McGuire et al. [32] later showed that the Y113F mutant in full length PGRMC1 binds heme. Kabe et al. [4] concluded that heme-mediated dimerization was required for the EGFR interaction. However, Y113 is also the phosphate acceptor of a membrane trafficking motif, and it is known to be phosphorylated in mammalian cells which would certainly prevent heme-binding [66]. Kabe et al. [4] looked for and observed no tyrosine phosphorylation of Y113, however such phosphorylation need only be transient (e.g., during transit from one location to another) and pertain only to a trace fraction of the bulk PGRMC1 population, and therefore the result was inconclusive. An alternative explanation offers itself, that EGFR interaction requires membrane trafficking, and does not involve heme-bound PGRMC1 (see Fig. 6A for schematic elaboration). Kabe, Suematsu and colleagues [92] provided additional evidence in favor of the existence of a dimer, or at least a higher order complex. They showed that the co-immunoprecipitation of endogenous PGRMC1 in a protein complex with a FLAG epitope-tagged PGRMC1 was prevented when cells were treated with succinyl acetone, an inhibitor of heme biosynthesis [88]. Therefore, the complex between endogenous and FLAG-tagged PGRMC1 was dependent upon the presence of heme. It remains formally possible that higher order complexes, rather than dimers, are formed between heme containing PGRMC1 monomers. However, the heme-stacking structure of the crystal structure [4] could indeed exist for PGRMC1 in mammalian cells. Furthermore, their recent publication showed that the saponin glycyrrhizin (major active anti-inflammatory and sweet-tasing molecule from liquorice root) and some derivatives bind to PGRMC1 residues that form the dimeric protein interface. The presence of glycyrrhizin caused enhanced deuterium exchange on those residues, indicating that their solvent exposure was increased by the dimer interface-disrupting molecule [92]. The presence of a regulated higher order complex in mammalian cells is also consistent with those results.
As discussed above, PGRMC1 could be involved in heme chaperoning [67, 85]. It exhibits reversible heme binding chemistry described above (Fig. 6A), and interacts with and regulates ferrochelatase, the final enzyme in heme biosynthesis [32, 85]. It has been proposed that ferrochelatase transfers heme to PGRMC1, which then transfers it to other proteins for distribution in the cell. This may include to PGRMC2 to transport heme to the nucleus [32, 67, 82, 85]. McGuire et al. [32] recently found that the PGRMC1:ferrochelatase interaction requires Y113, which interacts via a negatively charged tyrosinate group with the positively charged iron atom of heme. Contrary to expectations from a bacterially-expressed PGRMC1 Y113F protein with an N-terminal deletion (up to and including the first few residues of the MAPR domain) [4] which did not undergo heme-mediated dimerization, full length PGRMC1 Y113F mutant bound heme [32]. As mentioned above, the closely related PGRMC2 has recently been identified as a heme chaperone between the mitochondrial site of heme synthesis and the nucleus [82].
To extend this consideration, and strikingly for the thesis of this paper, cells
lacking PGRMC2-dependent heme-chaperoning exhibited dramatically impaired
mitochondrial function in brown fat cells, with double PGRMC1/2 knockouts showing
“similar, and perhaps greater, defects” [82]. This is consistent with the
hypothesized ancestral MAPR role in mitochondrial regulation. In the absence of
PGRMC2 and therefore nuclear heme, transcriptional repressors Rev-Erb
The heme-binding-deficient PGRMC2 protein used was achieved by mutating mouse Y131F (cognate with PGRMC1 Y107), K187A (cognate with PGRMC1 K163) and Y188F (cognate with PGRMC1 Y164: see Fig. 6B for human PGRMC1 numbering) to generate the 3xM triple mutant. PGRMC1 K163/Y164 contact heme propionate groups [4]. Importantly, this did not mutate the tyrosinate iron chelating PGRMC2 Y137 (cognate with PGRMC1 Y113) which is also potentially phosphorylated and involved in membrane trafficking [66].
While heme occupancy should certainly be impaired, it remains formally possible that these mutations could impair or enhance membrane trafficking since the mutated residues lie on opposite sides of the MIHIR in primary sequence and so the MIHIR forms a functional extension of the surface that binds heme. The here hypothesized MIHIR interactions with cytoskeletal components could be impaired if these require residues in the heme-binding pocket. Regardless of whether the effects are caused by altered heme binding (which does occur) or membrane trafficking (which is speculative), the unique heme-binding properties of PGRMC2 are important for heme trafficking to the nucleus in mammalian biology, and likely reflect properties at least partially overlapping with PGRMC1 through shared common inheritance.
We can here gather a few key observations from above. (1) The ancestral eukaryotic heme synthetic pathway came from the proto-mitochondrial alphaproteobacterial endosymbiont. (2) PGRMC1 interacts with and regulates ferrochelatase, the last and rate-limiting step of heme synthesis. (3) PGRMC2 shuttles heme to the nucleus to regulate genes that control mitochondrial activity and oxidative stress. Taken together, based on heme biology alone (i.e., independent of sterol biology and membrane trafficking) we could reasonably prosecute the case that the ancestral PGRMC/MAPR protein is hypothetically implicated in the process of eukaryogenesis, and the integration of mitochondrial and nuclear processes.
The effects of PGRMC1 on mitochondria, and its involvement in one of the most conserved eukaryote-specific reactions, prompted the author to explore a possible role in eukaryotic origins. We have recently discovered that MAPR proteins are related to a newly discovered subclass of prokaryotic cytb5 proteins, which we called cytb5M [8, 33]. These proteins that share structural features with MAPR proteins are found in diverse archaea and bacteria and are obviously descended from a very ancient ancestral gene. Hitherto described non-MAPR cytb5 domain proteins chelate heme via a two histidine (2-his) mechanism [86, 103]. Most cytb5M sequences indicated a 2-his mechanism by sequence alignment, and we confirmed this mechanism by solving the crystal structure for two cytb5M proteins [8, 33], albeit with altered orientation of heme binding relative to conventional cytb5 proteins. However, we also discovered a small subset of cytb5M-related proteins which shared with MAPR the tyrosines involved in the otherwise unique tyrosinate heme chelation of MAPR proteins (Fig. 6B,C). We designated this subclass of tyrosinate chelating proteins as cytb5MY (cytb5M with Y heme chelation) [8].
Strikingly, all cytb5MY proteins belonged to the candidate phyla radiation (CPR) bacteria [8] (Fig. 6C), a group of bacteria whose existence only relatively recently became apparent in the modern era of high throughput environmental sequencing. Although no living organism has ever been cultured, they are known from contiguous genome sequences discovered by high throughput environmental sequencing projects, and form a previously unappreciated major branch of life [104, 105]. CPR bacteria are typically obligate symbionts, characterized by reduced genome size, and often lacking genes required for usually essential pathways, such as (and possibly quite relevantly to our story) fatty acid synthesis [106, 107, 108].
It remains unclear how the membrane lipids of CPR are derived, since they cannot de novo synthesize them, and all biological membranes are synthesized in pre-existing membranes. This may be relevant for eukaryogenesis, as argued below, if a CPR lipid transfer system involving a cytb5MY protein was co-opted by the proto-eukaryote. It therefore appears quite possible that MAPR proteins entered the genome of the last eukaryotic common ancestor (LECA) from a symbiotic CPR bacterial genome. However, it remains possible that cytb5MY proteins are the result of horizontal gene transfer from a eukaryote with subsequent loss of the MIHIR region. Despite trying very hard, we could not resolve this issue [8]. The following discussion considers the implications if the former is the case, and CPR cytb5MY proteins were involved in eukaryogenesis.
Analysis of CPR genomes identified a consensus operon structure, which may have resembled the one involved in a putative eukaryogenic CPR bacterium (Fig. 6D). Known CPR cytb5MY genes are often found in operons containing cytb5MY, two other cytb5-related proteins (with 2-his heme chelation), a putative ferric reductase-like (pFre) gene, another gene of unknown function, a two-component inducible signal transduction unit, and possibly a gene for heme- and sterol-binding tryptophan-rich sensory protein (TSPO), also known as translocator protein, or peripheral benzodiazepine receptor (PBR). All proteins are transmembrane proteins. Since bacterial operons often encode proteins of related function, these genes in extant CPR genomes probably encode proteins that contribute to an inducible redox-related membrane complex [8]. It is tempting to speculate that the pFre enzyme alters cytb5MY heme iron oxidation to accept or jettison heme (Fig. 6A), however that remains speculative.
Nothing more is currently known about CPR cytb5MY proteins. However, we can reasonably speculate about at least one role played by cytb5MY if CPR were involved in the eukaryogenic symbiosis. From yeasts to mammals [31, 68] PGRMC-like proteins interact with CYP51A1 to demethylate lanosterol (Fig. 7A, Ref. [32]). CYP51A1 is the most strongly conserved eukaryotic cyP450 enzyme [39], suggesting that it serves a centrally important role in eukaryotic biology.
 Fig. 7.
Fig. 7.Proposed MAPR origins and CYP51A1-mediated steroidogenesis. A hypothetical model for the contribution of cytb5MY proteins to eukaryogenesis. In the presence of molecular oxygen, cytb5MY converted lanosterol to 14-dimethyl-14-dehydrolanosterol. Dissociation of heme provided a new binding site for the steroid product. Membrane-trafficking conceivably directed this to the endosymbiotic proto-mitochondrial bacteria to activate oxidative phosphorylation (Ox Phos) and detoxify oxygen. Membrane trafficking may originally have been related to the acquisition of membrane lipids by the CPR bacterium, since CPR often cannot synthesize fatty acids. This in turn may have contributed to the origin of the eukaryotic ester-linked membrane lipids as opposed to the ether-linked lipids of the original archaeal host cell. The acquisition of the MIHIR may have adapted the CPR protein to the archaeal host cell actomyosin system. Note that McGuire et al. [32] argue that the PGRMC1 heme iron atom is unlikely to act as an electron carrier in cyP450 reactions as depicted. However, it remains possible that regulation of heme iron oxidation state modulates PGRMC1 heme occupancy.
Lanosterol, the first steroid produced, is a five-ring organic molecule synthesized as a product of the mevalonate pathway (MVP). The triterpenoid squalene is formed by the condensation of six isoprene units by the MVP and cyclized into lanosterol involving the combined action of first squalene epoxidase and then oxidosqualene cyclase (also called lanosterol synthase), which produce a characteristic hydroxyl on the 3 position of sterols (Fig. 2). MVPs can be found in organisms from all kingdoms of life. In bacteria, squalene is also produced via the MVP and cyclized to six-ringed structures called hopanoids by squalene-hopene cyclase (SHC) enzymes that share distant homology with eukaryotic squalene cyclases [34, 109]. Lanosterol can be therefore regarded as an atypical 3-hydroxylated eukaryotic hopanoid that is largely produced by originally bacterially descended genes. The bacterium that most critically required regulation in our eukaryotic family tree was the proto-mitochondrion.
The synthesis of cholesterol from lanosterol produces diverse intermediary bioactive sterols (Fig. 3), which can each evoke their own biological responses fully independently of cholesterol production [36, 110]. Obviously, the very first of these reactions evolved first. It is the one involving lanosterol 14-demethylation catalyzed by PGRMC1 and CYP51A1 (Figs. 2,3). Our analysis showed that the required enzymes, including the genes for the MVP, squalene cyclase (which were previously known), CYP51A1 and PGRMC, all originated in bacterial rather than archaeal genomes [8]. Therefore, it is probable that a lanosterol-like hopanoid was demethylated by a cytb5MY/CYP51A1-like reaction in the symbiotic proto-eukaryote during the protoeukaryotic evolutionary response to molecular oxygen. (This has the status of reasonable speculation).
Interestingly in this context, oxygen levels regulate both the proteolytic activation, stability, and subsequent proteasomal degradation of the Sre1 homolog to mammalian SREBP proteins (which induce mevalonate pathway and steroidogenesis), whereby the MVP is upregulated under hypoxic conditions to compensate for lowered levels of oxygen-dependent steroid synthesis. Sre1 is destabilized in the presence of oxygen when sufficient steroids are available. Sterol synthesis in this system is oxygen-dependent, and also inhibits Sre1 proteolytic activation [111, 112]. Therefore, in yeast, steroidogenesis is regulated by oxygen, and occurs in the presence of oxygen.
It has long been noted that the squalene monooxygenase and CYP51A/PGRMC
reactions at the base of eukaryotic sterol synthesis both require molecular
oxygen (Fig. 2), which could mean that they first appeared as a response to
elevated levels of oxygen. However, 3-OH sterols appear in the fossil record
about a billion years before eukaryotes are thought to have arisen, and
alternative anaerobic reactions (e.g.,
Notwithstanding, and conceptually noted here for the first time, the combination of tyrosinate heme chelation and dissociation leading to altered MAPR states, as well as the dependence of the squalene monooxygenase and cytb5MY/CYP51A1 reactions on oxygen, potentially enabled a regulated response to oxygen by early eukaryotes (Fig. 2) (and the regulation of yeast SREBP pathway leading to MVP induction by oxygen levels [68, 112]). This would provide a potential mechanism to regulate the metabolic activity of the proto-mitochondrion. In this respect, it is striking that PGRMC2 regulates mitochondrial gene expression by reversible binding and shuttling of heme from the mitochondria (where heme is synthesized) to the nucleus (where genes encoding most mitochondrial proteins are located) [82] and PGRMC1 can regulate mitochondrial association with the ER [113].
Hopanoids lower the permeability of bacterial membranes to protons, which increases the efficiency of electron transport chains by increasing the proton gradient [34, 109]. Mitochondrial membranes require cholesterol to function, although elevated levels can be pathological [114]. A model emerges where genes from a symbiotic CPR bacterium were potentially involved in regulating the metabolic activity of the proto-mitochondrion in a symbiotic proto-eukaryotic cell collective by regulating sterol synthesis and transport (Fig. 7). This model is hypothetical and requires substantiation.
Because many CPR are unable to synthesize fatty acids, they must be able to somehow ferry membranes or at least lipids from host cell membranes. In this respect, the membrane trafficking role of MAPR proteins may have been involved in the eukaryotic origins of vesicle trafficking, or of lipid droplet formation. This model is even more conjectural and is also unsubstantiated.
This all implicates MAPR proteins with a previously unimagined (and still uncertain) role in the origins of eukaryotes. Strikingly, the products of the PGRMC1/CYP51A1 reaction in mammals are either follicular fluid meiosis-activating sterol (FF-MAS) or dihydro-FF-MAS (Figs. 2,3) [36, 110], which are both inducers of meiosis, a major innovation of the LECA relative to its prokaryotic forebears [115]. The association of PGRMC1 with the mitotic spindle kinetochore [57, 61] and its role in ensuring the ‘faithful progression through mitosis and meiosis’ [57] are therefore most interesting in this context (See also the review by Lodde et al. [116]). To better develop this theme and garner an appreciation for the importance of oxygen to eukaryotic origins, we should consider eukaryogenesis in closer detail. We (eukaryotes) are children of oxygen who would not have evolved in a world without photosynthesis.
The last common ancestor of cellular life was already living before the end of the heavy bombardment of the young earth by smaller bodies in the early solar system, more than 3900 million years ago (mya). The divergence of Eubacteria and Archaea occurred sometime after 3400 mya. Later, the eukaryogenic symbiosis between an Archaean host cell and a proto-mitochondrial alphaproteobacterial symbiont occurred later than 1840 mya, after the great oxidation event (GOE) that corresponded to the appearance of bacterial photosynthesis (photosystem II which generates molecular oxygen) [35, 117], which occurred rapidly at about 2330 mya [118]. The GOE has been proposed to have altered the levels of greenhouse gases such as methane to cause plummeting global temperatures in an event called the Makganyene Snowball Earth Event, which is the only certain low-latitude glaciation of the Palaeoproterozoic period [35]. Therefore, eukaryogenesis can be validly considered in terms of a response to the new threat of highly toxic molecular oxygen and changed environmental opportunities.
Eukaryotes are now considered phylogenetically to be a subgroup of the Archaea (analogously to English belonging to the Germanic language group, but also containing many words derived from Latin and other languages [119]). The archaeans with closest living relatives to eukaryotes are called the Asgard archaea. It is assumed that most medical biologists are unfamiliar with these areas of knowledge, and so they will be introduced briefly.
Like CPR bacteria, the Asgard archaea have until relatively recently been known only from their genomic sequences, obtained from projects that sequence the DNA present in different ecologically sampled environmental specimens. Asgard archaea were first discovered as part of the deep-sea biota at a site called Loki’s castle in the North Atlantic, and subsequent clade members have all acquired nomenclature related to the Viking pantheon. The Lokiarchaeota are associated with deep sea mid-oceanic ridges, where tectonic plate movements are caused by superheated lava welling up to the surface of the ocean bed (for reviews: [120, 121]).
Phylogenomic analysis of the Asgard archaea suggests that the proto-eukaryotic host cell may have been mixotrophic, with a facultative aerobic metabolism [122]. Mixotrophs obtain energy in an intermediate state between autotrophy (e.g., photosynthesis) and heterotrophy (consuming organic molecules from other organisms), and mixotrophy often involves biotrophic symbioses [123].
The reconstructed host cell possessed its own complete glycolytic pathway and tricarboxylic acid cycle (TCA). The presence of rhodopsins in the most closely related Heimdallarchaeota Asgard group (the closest archaeal relative to eukaryotes discovered to date) suggests that the host cell may have at least transiently occupied the upper oxygen-enriched ocean layers [120, 122, 124]. Asgard DNA appears to be spatially segregated from ribosomes [125], suggesting that membrane compartmentalization and perhaps a nucleus may have already existed in the Asgard proto-eukaryotic host cell. PGRMC-mediated heme shuttling to the nucleus to regulate mitochondrial activity (see section 2.1.3 above) may therefore reflect a function that could have contributed to eukaryogenesis.
The presence of a TCA cycle in Asgard Archaea, and therefore the archaeal host, clearly indicates that the selective advantage of mitochondria was not related to the increased ATP yield of the TCA cycle over glycolytic metabolism. Rather, the selective advantage of the proto-mitochondrial TCA cycle to the symbiotic chimera may have been that the final electron acceptor of the proto-mitochondrial electron transport chain was oxygen because of a mutation that enabled an ancestral alphaproteobacterium to survive in an oxygenated environment: a triviality that may have spawned the world as we know it.
For a discussion of the recently proposed “molecule-unbound ion-radical” (murburn) theory of the origins of mitochondrial oxidative phosphorylation, see Manoj and Bazhin [126]. In this model, oxidative phosphorylation originated from a series of redox reactions that deal with diffusible reactive oxygen species. Relevant here, the properties of molecular oxygen as final electron acceptor are proposed to have enabled quicker metabolic responses to environmental stimuli by murburn-centered life.
The majority of genes of the LECA appear to have arisen de novo during the process of eukaryogenesis, being unrecognizable in the ancestral Asgard archaeal or alphaproteobacterial proto-mitochondrial groups [121]. Some of these may of course have been acquired from now extinct prokaryotic lineages, or yet uncharacterized ones. Strong evidence also exists for contributions to the eukaryotic genome via horizontal gene transfer from other broadly identifiable bacterial groups during eukaryogenesis [121]. Indeed, it has been proposed that the mitochondrial acquisition was a relatively late event in eukaryogenesis, with the archaeal host cell genome already being chimeric, having acquired many genes including some associated with signaling, metabolism, nucleus, and both plasma- and endo-membrane biology from several different recognizable non-alphaproteobacterial bacterial groups. The named groups did not include CPR bacteria [127].
In further comparative genomics and phylogenetics study Spang et al. [124] also reconstructed the archaeal host cell as an organoheterotroph (i.e., which gained sustenance from the organic molecules of other organisms) that provided hydrogen or another high energy electron source to a symbiont separate from the proto-mitochondria. This symbiont was proposed to have acted as an electron sink in anoxic or micro-oxic environments [124]. Ettema’s group has recently identified that the actin cytoskeleton (relevant to PGRMC membrane trafficking) arose from the archaeal host cell [128], and that several components of eukaryotic hydrogen metabolism were inherited from an anaerobic member of the chlamydiae group of bacteria, which they proposed contributed to an evolutionarily mosaic origin of eukaryotes [129].
Imachi et al. [130] describe the first culturing of a live Asgard
archaean species, called MK-D1, of the Lokiarchaeaota clade. It was isolated from
a sediment core of a mid-oceanic ridge environment and cultured under anoxic
culture conditions in the presence of methane, CO
Imachi et al. [130] reconstruct a eukaryogenic scenario in the hyper-oxygenated post GOE world, where organic matter was produced mostly by photosynthetic bacteria in the upper, now oxygenated, levels of the ocean. Sulfate concentrations became elevated [118] (literature citations in this paragraph are those of Imachi et al. [130]), which favored syntrophy with sulfate-reducing bacteria [132]. In this world most organic material was in the upper ocean levels, however excess sediment fell to benthic dwellers of the anoxic depths. It was proposed that to move upwards and exploit the abundant organic resources available, the proto-eukaryotic Asgard archael host cell entered symbiosis with its alpha-proteobacterial symbiont, and its oxygen-accepting electron transport chain [130]. Thereby the facultative aerobic capacity of the host cell could be switched, to dramatically consume toxic oxygen by feeding oxoacid carbon skeletons to its endosymbiont in the presence of high oxygen levels, or it could metabolize those same amino acids to generate hydrogen for a sulfur-reducing syntrophic partner under lower oxygen tension. In this scenario eukaryogenesis was not coincident with the GOE, but occurred later to deal with the GOE by enabling access to the energy-rich oxygenated surface waters as available habitat [130]. In support, based upon the presence of suites of genes associated with various modes of motility in different eukaryotes, it has been suggested that the LECA may have been able to switch between ameboid crawling (i.e., suitable for deep sea floor benthic life) and flagellated motility (i.e., suitable for life in the oxygenated surface waters) [133, 134].
However, it occurred, the facultative switch between anaerobic metabolic states and aerobic mitochondrial activity was driven by an interplay between hypoxia and hyperoxia that became genetically hardwired from the genomes of disparate organisms to enable survival of the syntrophic whole. As genetic material was transferred to a central chimeric genome, some of the pre-existing circuitry is likely to have remained intact if the emergent eukaryotic biology depended upon it. Similarly, new genomic environments may have led to new functions for some genes/proteins.
This may have (perhaps) involved the putative ancestral CPR-derived MAPR protein. Under this model, which could conceivably be still hardwired into our modern genomes, the hypoxic response and regulation of mitochondrial activity becomes a switch between deciding to feed the mitochondria and consume oxygen, or to produce reducing equivalents and feed the sulfate-reducing syntroph (a CPR bacterium?), in which case the carbon skeleton could not be catabolized in the mitochondria.
In terms of this review, we can view autophagy as a system for generating non-glucose carbon skeletons for metabolic energy production, and hence we can place autophagy near the metabolic functional heuristic core of PGRMC1’s proposed ancestral eukaryogenic role (regulating energy production by mitochondria in response to oxygen levels). Autophagy plays a critical role in the metabolism and clinical phenotype of many tumors [135, 136], and other pathologies such as Alzheimer’s disease [137]. PGRMC1 regulates the induction autophagy by forming protein complexes with MAP1LC3 (microtubule-associated protein 1 light chain 3, or LC3) and UVRAG (UV radiation resistance associated/UV radiation associated gene) [138].
PGRMC1-dependent autophagy can be pharmacologically activated to metabolically
render cultured ovarian cancer cells more likely to undergo apoptosis, and more
sensitive to cis-platin treatment [139]. PGRMC1 knockdown promotes the
differentiation of human pluripotential stem cells coincidentally with a
reduction in autophagy and elevated activity of the p53 and Wnt/
Organizer activity requires Wnt/
It has been proposed that (at least part of) PGRMC1’s P4-responsiveness may be
ascribable to interactions with PAQR proteins [20, 47]. With respect to
eukaryotic origins, it is notable that whereas most G-coupled protein receptors
originated from the archaeal host cell, the PAQR-type proteins are of bacterial
origin [148]. It is therefore conceivable that a MAPR/PAQR-GPCR response was
involved in metabolic regulation of proto-mitochondrial metabolism. In this
context it is also noteworthy that the PGRMC1-dependent P4 induction of
glycolytic Warburg metabolism in HEK293 cells involves G
That such immediate-early gene induction of anabolic metabolic phenomena by PGRMC1 may be widespread is consistent with observations e.g., from neural progenitor cells that P4 increases cell cycle gene expression and proliferation in a PGRMC1/2-dependent manner [58]. Therefore, autophagy may be inversely related to active mitogenic signaling, suppressing Wnt signaling while the cell hunkers down to inhibit cell division until environmental nutrient scarcity is overcome. Direct regulatory modulation of the Wnt and autophagy pathways by PGRMC1 is consistent with the ancient eukaryotic role of PGRMC1 proposed in this work and may be related to the metabolic control of the Warburg Effect.
A MAPR-regulated control of metabolism via the autophagic supply of amino acid fuel supply may be ancient in eukaryotes, since the plant MAPR protein MSBP1 (Membrane Steroid Binding Protein 1) associated with starvation-induced components of the reticulophagy (the selective autophagy of ER proteins) system [153]. The MAPR protein was identified as a substrate for autophagy in this plant system. It would be interesting to assay whether reticulophagy would operate without MSBP1, if indeed autophagy is part of an ancient MAPR-dependent mechanism of metabolic regulation related to eukaryogenesis.
A main theme of this present paper, extending thoughts from a previous publication [8], is to consider the implications if the ancestral CPR ctyb5MY protein that gave rise to MAPR proteins like PGRMC was indeed involved in the regulation of mitochondrial activity and cell metabolism in the LECA (which may or may not be true). In any case, a relatively early eukaryote must have acquired a MAPR gene. It may have come from an inducible CPR operon (which contained a two-component element) that contained a putative ferric reductase and three cytb5-domain proteins: the cytb5MY gene and two others which are unrelated to eukaryotic genes (Fig. 6D). Only MAPR proteins are identifiably related among eukaryotic proteins to this consensus CPR operon. However, the operon in several CPR species also contains the TSPO gene [8], suggesting possible functional association between MAPR proteins and TSPO (Whether eukaryotic TSPO is descended from CPR bacteria was not examined in detail, but CPR TSPO proteins do not give the highest BLAST scores compared to other bacteria using human TSPO as search query, and therefore CPR TSPO is unlikely to have been ancestral to the eukaryotic proteins).
TSPO proteins are highly evolutionarily conserved, being found in eukaryotes, bacteria and archaea [154, 155]. Like PGRMC1, mammalian TSPO’s functions remain elusive. It is observed at several different subcellular locations: primarily the mitochondrial outer membrane, but also the cytoplasmic membrane as well as nuclear and perinuclear localization following stresses such as injury or inflammation [156, 157]. Its natural ligands are suggested to include heme, cholesterol, and the protein called Diazepam Binding Inhibitor (DBI) (also known as Acyl-CoA-binding protein), an acyl-CoA ester-binding protein [158]. However, the physiological relevance of identified ligands remains unclear [155]. TSPO plays prominent roles in oxidative stress response and inflammation and has been strongly associated with steroidogenic tissues (for reviews, see [154, 158]).
Earlier literature associated TSPO with steroidogenesis and cholesterol transport into the mitochondria, however it has now become clear that despite high expression in steroidogenic cells, TSPO is not required for steroidogenesis [159]. It does appear linked to mitochondrial fatty acid oxidation [160]. Hiser et al. [154] propose that TSPO was originally a porphyrin-binding bacterial stress protein, which has gained new functions in the course of evolution. Its contributions to redox and inflammatory homeostasis [154] are consistent with that conclusion. If an original steroidogenic cytb5MY protein was co-expressed in a CPR operon with a TSPO allele [8], then TSPO and PGRMC1 may jointly or reciprocally modulate some ancient eukaryotic processes.
Lee et al. [161] show that PGRMC1 is involved in a pro-inflammatory response involving EGFR in hepatocellular carcinoma. It will be interesting to see whether TSPO is involved in that system. Considering the involvement of PGRMC1 in regulating fatty acid metabolism [41, 43, 162, 163, 164, 165], and PGRMC1’s profound effects on mitochondria [50, 113, 165], and its association with stress response [96, 97, 98, 99], the overlap between TSPO and PGRMC1 biology is extensive, and fully consistent with an ancestral relationship. It is tempting to speculate that a cytb5MY protein might have transferred its heme to TSPO in a redox-dependent manner, thereby changing the responsiveness of cytb5MY and TSPO.
In preliminary work we show that TSPO is required for sigma-2 receptor (S2R) activity of TMEM97 in at least MIA PaCa-2 pancreatic cancer cell binding of the fluorescent SW-120 S2R ligand [166]. TMEM97 and PGRMC1 form a protein complex that is involved in membrane trafficking [149, 150, 167], and like PGRMC1, TMEM97 is implicated in sterol biology (for review: [168]). Recently 20(S)-hydroxycholesterol has been shown to be an endogenous ligand of TMEM97 [45]. We did not observe interaction between PGRMC1 and TSPO, however, both TSPO and PGRMC1 interacted with TMEM97 by proximity ligation assay [166]. Those preliminary results are currently being investigated further. The CPR operon structure is clearly involved in some inducible redox process, and the cytb5MY gene would have been of such central importance to eukaryogenesis that it could not be lost to eukaryotes.
One of the common features of eukaryotic cells from that time to this is the mitochondrion. PGRMC has coevolved with several classes of mitochondrial genes [67], and PGRMC1 phosphorylation status imposes quite dramatic changes on the way mitochondria behave [50], as if a spanner had been thrown into the mechanism of this ancient system. As intriguing and plausible as this hypothesis is, it clearly requires further substantiating validation.
The requirement of the PGRMC/CYP51A1 most highly conserved eukaryotic cyP450 reaction for molecular oxygen (Fig. 7A) is extremely relevant in this context. It allows us to develop a hypothetical scenario where in the presence of oxygen, lanosterol is demethylated to produce follicular fluid meiosis-activating sterol (FF-MAS). The resulting reduced heme would dissociate from PGRMC, potentially creating a binding site for the newly formed FF-MAS (Fig. 7B). This conceivably also facilitates a newly acquired eukaryotic cytb5MY membrane trafficking function via the MIHIR, which is not present in known CPR cytb5MY genes but was acquired by the first eukaryotic MAPR gene (whether or not CPR were involved) (Fig. 6B,C). This is all speculative.
Interestingly most CPR cytb5MY contain two glycine residues at the point where the MIHIR is inserted (Fig. 6C). The GG sequence would enable a sharp turn in the polypeptide backbone between the two helices present in the canonical cytb5 fold [8]. This corresponds in the sequence alignment of Fig. 6C to PGRMC1 C129/L130 which mediate direct hydrophobic contacts with heme [4]. Insertion of the MIHIR sequence between these two helices, at a point of heme contact (Fig. 6C), was the historical original defining feature of the MAPR family [1], in which those two glycines are not conserved. It would be surprising if MIHIR function and heme occupancy were not interrelated.
The proto-MAPR protein may have transported a sterol/hopanoid to the mitochondrial membrane to activate the TCA cycle in the presence of oxygen (Fig. 7). The sterol-associated biology involved still seems to be mirrored in modern PGRMC1 biology [47, 67, 150]. Heme chaperoning or regulation of heme synthesis [67, 82, 85, 169] may also have been involved. This, of course, is hypothetical speculation.
Cytoplasmic membrane trafficking itself, where vesicles are pinched into the cytoplasm in an actin-dependent process, was thought to have arisen newly in eukaryotes [170], however genes for both a dynamic actin cytoskeleton and cytoplasmic vesicle transport are widespread throughout the Asgard archaea [128]. Motivated from this perspective, we recently identified that the PGRMC1 MIHIR contains a predicted coiled-coil protein interaction motif that is like motifs in many myosin proteins [7]. A coiled coil is a protein interaction motif where alpha-helices from two protein interact with each other via hydrophobic and other interactions between two helices. Because of the helical register of residues, aliphatic residues are spaced with a heptad repeat pattern in the sequence [171].
Myosins are the motor proteins associated with the actin cytoskeleton, and the implication is that PGRMC1 interacts with some of the same proteins as myosins. We [91] and others [172] have discovered that PGRMC1 is present in protein complexes with mitochondrial proteins, and with components of the actin cytoskeleton. The latter interactions are sensitive to the small molecule inhibitor AG-205 [91], which was designed to occupy the MAPR heme-binding site [173] (But see Section 2.1.1 above.)
Considering the above, the new eukaryotic acquisition of the MAPR MIHIR was conceivably related to eukaryogenesis: perhaps originally involving CPR membrane exchange because many CPR bacteria cannot synthesize their own fatty acids, and the MIHIR was probably required in the LECA. The hypothesis here is that MAPR membrane trafficking was involved in supplying MAPR-synthesized sterols and/or perhaps heme to regulate the mitochondrion in the presence of oxygen. It has been observed that eukaryogenesis must have been an inherently improbable event, having arisen in this solar system with the same frequency as life itself (once each) [170]. Perhaps the unique cytb5MY with a membrane-trafficking MIHIR was instrumentally enabling in the process. Clearly, this field merits further research.
Hydrophobic heptad repeat residues of the predicted coiled-coil region of the MIHIR (Fig. 8, Ref. [7, 53, 174, 175, 176, 177, 178, 179]) are conserved in MAPR proteins (although predicted coiled-coil formation is not conserved) [7], and appear to be inherited from the ancestral eukaryotic MAPR MIHIR sequence. This was inserted into the MAPR cytb5 domain fold in the middle of the heme-binding domain. Heme interacting Y107 (H-bond) and Y113 (tyrosinate heme chelation), and multiple hydrophobic interactions with heme (G124, L125, F128, C129 and L130) are N-terminal to the MIHIR, whereas propionate H-bond forming K163 and Y164 are C-terminal to the MIHIR (Fig. 6C) [4, 180]. We can then imagine that the MIHIR, or proteins interacting with it, could exert strong allosteric influence over heme affinity, and probably vice-versa. This interrelationship is here hypothesized to have been critical in eukaryogenesis, a process which involved the host cell and mitochondria adapting to each other permanently.
 Fig. 8.
Fig. 8.Early animal evolution of PGRMC. (A) The basic evolutionary tree topology depicts a simplified version of that presented by Sperling and Stockey, who combined paleontological, organic, geochemical and molecular clock evidence [174]. The depicted branch times in millions of years ago represent approximate averages without 95% confidence intervals from Sperling and Stockey. See also Gold [175]. Placozoans and ctenophores were absent from Sperling and Stockey. Placozoans are placed according to the acquisition of Hif-1 [53] and shared gene duplications [176], and ctenophores are placed as a metazoan outgroup following Erives and Fritzsch [176, 177]. The bracket (?) joining the last eumetazoan common ancestor (LEUMCA) and last eumetazoan and placozoan common ancestor (LEPCA) acknowledges the possibility that placozoans are descended from the LEUMCA [178, 179] (i.e., previously underwent gastrulation which they have secondarily lost). Vertical shaded bars represent major Neoproterozoic glaciations. The first animal fossils are known from late Ediacaran rocks. See Sperling and Stockey for details. The relative tree topology for the branch between ctenophores and other metazoans is presumed to be correct. However, the indicated time of this branch is solely estimated by the author. (B) PGRMC evolution in pre-Ediacaran fauna animals. Protozoans and early-branching animals (EBA, ctenophores and poriferans) inherited a PGRMC gene with transmembrane helix (TMH) and cytochrome b5 MAPR domain that presumably reflects the ancestral condition of the last eukaryotic common ancestor. LEPCA has a C-terminal extension which was larger in the LEUMCA. The LEUMCA also acquired Y139 and Y180. Bilaterians additionally acquired presumed regulatory phosphorylation sites at T178 and S181. The N-termini are depicted dotted lines as these may exhibit non-systematic length variations between and within the indicated phyla. For source information, see [7].
Peluso and Pru [181] report that PGRMC1 associates with eukaryotic ribosomal translation initiation factors. In a pilot co-IP and proteomic protein identification experiment, Sarah Teakel in the author’s lab had also identified ribosomal proteins in the co-IP pellets using HA-tagged PGRMC1 as bait. For reasons explained elsewhere, due to lack of resources we were unable to generate replicate numbers to provide data of publication quality, and so were unable to pursue this. We also already had the proteomics pathways analysis that was later published in 2020 [50], suggesting that PGRMC1 phosphorylation status could affect the abundance of some proteins involved in translation, but also that the dramatic changes in protein abundance profiles that we observed could be due to alterations in the subset of cellular mRNAs being translated. That prompted the author to investigate hints of ribosomal association from published resources, where The Human Protein Atlas reported prominent localization of PGRMC1 to the nucleolus, the site of ribosome biogenesis, and speculate that PGRMC1 “may affect the composition of ribosomes, translation initiation factors” [47], as cited by Peluso and Pru [181]. Terzaghi et al. [182] later reported the nucleolar localization (i.e., the site of ribosomal RNA synthesis and pre-ribosomal particle assembly [183]) of PGRMC1 in bovine granulosa cells and oocytes.
We later discovered that PGRMC1 was inducing dramatic epigenetic changes in the MIA PaCa-2 cells we worked on, which could have been sufficient to induce the observed large changes in protein expression without the need to invoke effects on ribosomes and selective mRNA translation. However, the possibility that PGRMC1 can affect protein expression at the translational level gains credence from the report of Peluso and Pru. Like our lab, they had also been investigating PGRMC1-ribosomal interactions prior to 2016, and they recently reported the presence of several ribosomal elongation initiation factors in PGRMC1and PGRMC1 co-IP pellets [181]. To consider just one example, for PGRMC1 these included the Eukaryotic Initiation Factor 3 (EIF3) subunit EIF3B. The multi-component EIF3 complex binds to and regulates the translational efficiency of specific mRNAs involved in growth control, cell cycle, differentiation, and viability [184]. Its activity can be regulated by methylation via EEF1AKNMT (eEF1A lysine and N-terminal methyltransferase), which can affect the subset of mRNAs being translated and tumorigenicity [185, 186]. Therefore, PGRMC1 could possibly modulate the efficiency with which specific EIF3B-depdendent mRNAs (for example) are translated. This concept extends to other ribosomal factors.
Having spent many years working in the field of proteomics, the author experienced first-hand the developing awareness that mRNA levels are surprisingly poorly correlated with protein levels. It is now an accepted phenomenon that many mRNAs are only translated under appropriate regulated circumstances [187, 188, 189, 190, 191].
This point is expounded here in detail because this biology could well represent vestiges of an ancestral cytb5MY/MAPR role from eukaryogenic times, where modulation of host cell metabolism by an obligate symbiotic CPR bacterium may have been mediated via a cytb5MY/MAPR protein which was present in the host cytoplasm. This hypothesis predicts that PGRMC (and there is no reason to assume that either modern PGRMC1 or PGRMC2 functions are more typical of an ancestral state) modulation of ribosomal function would probably have initially served to alter host cell metabolism to provide metabolites for the obligate CPR symbiote. Such a switch may have controlled feeding carbon skeletons to either oxidative phosphorylation (mitochondria) or cytoplasmic lipid or amino acid syntheses. As such, PGRMC modulation of ribosomal function would be consistent with the hypothetical eukaryogenic role of MAPR proteins proposed in this current work. If this is correct then PGRMC1 modulation of ribosomal activity is predicted to modulate metabolism via regulation of mitochondrial function, and perhaps even cell cycle control if that is inherited from early eukaryotes. Please note that (1) the involvement of PGRMC1 in ribosomal translation remains formally unproven, and (2) even such proof would not lead to automatic acceptance of the eukaryogenic hypothesis. This avenue requires further investigation.
If the above scenario is correct then the MAPR proteins are highly conserved in eukaryotes because they mediate processes that were central to eukaryogenesis, and are still required by many eukaryotes, including animals. However, very little is known about Neudesin and Neuferricin protein functions in animal evolution beyond their phylogenetic distribution and neural roles [7], and therefore this paper focusses on the much better characterized PGRMC family.
It has recently emerged that evolutionary innovation may have once more resorted to tweaking PGRMC biology to enable the rise of eumetazoan animals. Eumetazoans are the group of animals that possess distinct adult cell types as a result of induced differentiation associated with a gastrulation organizer and include cnidarians and bilaterian animals [192]. To understand any role of MAPR proteins in animal evolution we must first understand the basic principles underlying the early evolution of animals, which the author once more assumes are unfamiliar to most medical researchers, and which are accordingly introduced in the following.
As time passed after the appearance of the LECA, eukaryotes diversified into multiple lineages, one of which gave rise to both fungi and humans. That lineage is called the opisthokonts, of which a subgroup called the holozoans became sophisticated predators able to engulf bacteria and feed off the cytoplasmic contents of eukaryotes as large as themselves [193]. This group developed a toolkit of new genes that would prove essential for subsequent animal evolution, importantly including the evolution of both tyrosine kinases, and SH2 domains that bind to phosphorylated tyrosines, which are absent from fungi but present in animals and their close protozoan relatives such as the Choanoflagellates (the sister group to animals) [73].
Choanoflagellates are the closest single-celled protozoans to animals, and multicellular animals share a common ancestor with that group [73, 194, 195]. Some choanoflagellates can form multicellular spheres, which are topologically like early animal embryos of a single cell layer sphere. There is superficial resemblance of the body organization of sponges or placozoans to multicellular choanoflagellates, featuring a layer of polarized flagellated cells that obtain sustenance from the exterior and achieve complex surface topologies via adherens junction-mediated movements by coordinated action of myosin-motored actin cytoskeleton (actomyosin) and adherens junctions [195]. However, a fundamental difference in body topology is that planar coalescences (sheets) of separate coalesced choanoflagellate cells can actively form a sphere. By contrast, the spherical blastula of all animals arises from the serial divisions of a single zygote cell, which is never observed in choanoflagellates [196].
The first animals evolved from a common ancestor with choanoflagellates sometime between perhaps 950 and 750 million years ago (mya) [197, 198], perhaps close to 800 mya (Fig. 8A), which coincided with the appearance of a suite of new metazoan-specific genes [197]. This was a time of low organic productivity, which was subsequently reduced even further by the approximately 75 million-year Sturtian glaciation event: the most prominent of the Neoproterozoic “Snowball Earth” glaciations thought to have frozen the world’s oceans. See also Gold for further consideration of this topic [175].
The Sturtian glaciation led to dramatically reduced bioproductivity, which corresponded to both reduced organic biomass availability, and even lower oceanic oxygen levels [174]. If the gastrulation organizer arose as a response to variable oxygen conditions, that could be retained in modern early embryological processes. Intriguingly, pluripotential embryonic stem cells (PSCs) exhibit a plastic metabolic state that can reversibly adapt to fluctuations in oxygen concentration. The most conspicuous two states have been described as “naïve” and “primed” PSCs. Naïve cells arise shortly after the zygote begins transcribing its genome, which rapidly becomes hypomethylated to remove parental epigenetic status. This involves net low methylation levels of genomic CpG and the repressive H3K27me3 (trimethylation at lysine 27 of histone H3). Naïve PSCs are found in the inner cell mass of the early blastocyst, which emerges with gastrulation, and can interconvertibly obtain ATP via either anaerobic glycolysis or aerobic oxidative phosphorylation (a metabolic switch that was implicated PGRMC1 with above).
During embryology, naïve cells later develop into primed PSCs, which in the mouse exhibit predominantly glycolytic metabolism, as well as hypermethylation of CpG sites and the repressive combination of histone H3K27me3 and H3K9me2/3 (di-methylation at lysine 9 of histone H3) methylation. Certain enhancer and promoter regions remain hypomethylated. In culture, the naïve-primed conversion is reversible, depending upon culture conditions [199, 200, 201, 202, 203], which include switching from 5% to 20% oxygen [204]. Accordingly, the implantation stage mammalian blastocyst is almost exclusively glycolytic [205, 206]. These are the developmental processes which give rise to the gastrulation organizer, and which probably somehow reflect its evolution to produce the LEUMCA, and perhaps the environmental adaptations required at that time.
If Sperling and Stockey’s estimated chronological calibration of major metazoan lineage divergences is correct, then the LEUMCA common ancestor of bilaterians and cnidarians arose perhaps just over 700 mya, corresponding roughly to the origin of the Sturtian glaciation (Fig. 8A). However, this figure represents the consensus from several different studies, each of which was associated with larger 95% confidence intervals. See the original work for details [174].
It is possible that multicellularity arose with the ancestor of choanoflagellates, or even earlier, and that modern metazoans had multiple choanoflagellate-like origins. However, it is also possible and perhaps more likely that animal multicellularity arose with an “Urmetazoan” common ancestor of poriferans, ctenophores, placozoans, cnidarians and bilaterians [194].
Much of the uncertainty has concerned the position of the ambiguous group known as ctenophores, which have a gut, mesoderm-derived muscle and a nervous system with synapses [207]. Placozoans and poriferans do not, although poriferans do contain many genes related to synapses (as indeed do the choanoflagellate single celled animal sister group) [208]. This is important when considering the origins of complex animal bodies, and especially nerves, in our direct lineage.
Ctenophores have ectodermal and endodermal nerve nets, but lack many synaptic genes present in the LEUMCA clade. It has long been controversial whether nerves evolved separately in LEUMCA and ctenophores. However, ctenophore nerves are specified late in development and related to ectodermal cells, in contrast to cnidarians where cells of the nerve net differentiate early in development from both ectodermal and endodermal cells [207].
Ctenophore early embryonic signaling molecules are also different to those of eumetazoans in terms of expression patterns and chemical identity. Furthermore, many traditional bilaterian neuronal markers are shared with cnidarians but absent from ctenophores. Like cnidarians, ctenophores have tentacles, a gut, nerves, and polarized epithelial cells. Their comparative embryological development, based upon comparison of different genetic pathways in the ctenophores than in cnidarians and bilaterians, provides very strong evidence for independent evolution of nerves (and therefore gut and muscle) in the LEUMCA and ctenophores. The complement of genes in the genetic toolkit of ctenophores is also more like poriferans than any other animal group. For such reasons, many recent authors have concluded that ctenophores are a sister group to all other metazoans [74, 177, 179, 194, 195, 209, 210, 211, 212, 213]. Ctenophores could even represent a separate evolution of multicellularity from a different choanoflagellate-like group than other metazoans [177]. From our own work, based only on MAPR proteins, the NEUFC C-terminus suggests that ctenophores share derived features with poriferans and other animals relative to at least the choanoflagellates we surveyed, while the PGRMC C-terminus supports a common ancestor of placozoans and eumetazoans but not poriferans or ctenophores [7], supporting the tree topology of Fig. 8A (albeit on the basis of just two genes).
Erives and Fritzsch [176] offer compelling evidence, based on shared inheritance of paralogous duplicated genes, that ctenophores indeed do form a sister group to all other metazoans, which has been followed in Fig. 8A. For that reason, ctenophores are not further considered in this paper, which assumes that the first appearance of nerves in the human lineage was in the LEUMCA. However, note that while it seems to be accepted that ctenophores are not descended from the LEUMCA, it remains debated by some whether an ancestral animal may have evolved nerves with subsequent loss by poriferans and placozoans [211].
The major animal multicellularity innovation was achieved prior to the LEUMCA: cadherin-mediated adhesion of cells to form an epithelial layer attached basally to a strong and flexible basal lamina enabled directed cadherin dependent and adherens junction-mediated shape changes by the actin cytoskeleton. This enabled the epithelial layer to undergo morphogenesis from an ancestral spherical shape into convoluted cup or tubular forms, as evidenced by early metazoan body grades best represented today by poriferans and placozoans [195, 214]. Cadherins themselves appeared before metazoans, however metazoan evolution was accompanied by the appearance of metazoan-specific genes with regulatory features on the cytoplasmic tails, as well as a small number of accessory proteins, to interact with them [215].
The LEUMCA developed new actin innovations. The type of muscle possessed by cnidarians permits us to reconstruct the probable LEUMCA situation. Although cnidarians vary considerably between genera, their body consists of two cell layers of primarily epithelial cells, with some ganglion neurons, sensory neurons, gland cells, and other specialized cnidarian cell types including nematocyte and nematoblast cells (involved in toxin delivery). Cnidarian muscle cells are epitheliomuscular, meaning they have both epithelial and contractile character. Perpendicular networks of ring muscle and longitudinal muscle fibers connect the adherens junctions of adjacent epithelial cells, which enables peristaltic motion (e.g., the swimming action of a jellyfish) [216, 217].
The novel cadherins described above led to regulatable adherens junction
activity in eumetazoans, adding new function to a pre-existing ancient machinery
that connected to the actin cytoskeleton via
 Fig. 9.
Fig. 9.A mammalian adhesion junction. (A) Sheets of epithelial cells
are connected by an actin-linked network of adherens junctions. (B) The schematic
organization of a mammalian adherens junction, showing the principal structural
proteins. Actin filaments are associated with adherens junctions in addition to
several other actin-binding proteins such as the F actin cross-linking protein
alpha actinin and the F-actin and E-cadherin adapter protein vinculin. The
vinculin head domain associates with E-cadherin via
Let us revisit consideration of the MIHIR, which appeared in the first MAPR protein early in eukaryotic evolution as discussed above. Early animal acquisition of Y139 in the MIHIR [7] may have contributed to eumetazoan evolution. It has been speculated but not demonstrated that the MIHIR may be involved in the attested membrane trafficking ability of PGRMC1 (for review: [47]). PGRMC1 is known to interact with components of the actin cytoskeleton [91, 172], and a portion of the MIHIR exhibits both strong evolutionary conservation and predicted propensity to form a coiled-coil protein interaction involving heptad repeat L residues discussed above (Fig. 10, Ref. [7]).
 Fig. 10.
Fig. 10.The PGRMC1 MIHIR contains a conserved amphipathic helix. (A) PGRMC1 showing the residues from the MIHIR that exhibit predicted coiled-coil propensity [7]. Asterisked residues form the predicted heptad repeat, while residues indicated by triangles are within 3 of a heptad repeat. The green triangle is Y139. (B) A LOGO plot of MIHIR residues from 20 bilaterians [7], showing levels of evolutionary conservation. A single large letter denotes only that residue at all sequences. The predicted region of coiled-coil formation [7] is indicated by a box. The black bar shows residues plotted in C. Other annotation follows A. (C) The 11 conserved MIHIR residues from B form an amphipathic helix with a negatively charged surface when plotted on a helical conformation.
That region is part of a motif which resembles similar motifs from the coiled-coil regions of multiple myosins [7], and would present a conserved negatively charged amphipathic helical surface (Fig. 10C). It is conceivable that an amphipathic helix is somehow involved in PGRMC1’s membrane trafficking function, however reported membrane trafficking proteins with amphipathic helices contain positively charged, or mixed positively and negatively charged helices, where the positive charges interact with negatively charged membrane lipids [218, 219]. It seems therefore more likely that this PGRMC1 putative amphipathic helix interacts with positively charged proteins (such as those involved in membrane trafficking?) that can also interact with selected myosins sharing the similar motif [7]. Thereby PGRMC1 would compete with myosin motor proteins for binding to some proteins (which could also regulate motive force required for membrane trafficking).
Notably, PGRMC1 Y139 would be one of the aliphatic residues that would contribute to coiled-coil inter-helical protein interaction surface (Fig. 9). This residue appears to have been a tryptophan in the single celled ancestors of animals, but to have mutated to a tyrosine by the time of the LEUMCA [7]. Both tryptophan and tyrosine, being large hydropathic residues, should be compatible with a hydrophobic surface involved in coiled-coil formation. However, the conserved aliphatic helical surface of Fig. 9C would be destroyed by phosphorylation of Y139. This would (1) interrupt any coiled-coil interaction, and (2) facilitate new interactions with SH2 domain proteins [7] (Fig. 8B).
Therefore, we can imagine that the LEUMCA could retain whatever functions were performed by the predicted ancestral hydrophobic helical surface, but that it could switch these off by Y139 phosphorylation. Thereby, novel functions associated with the recruitment of SH2 domain proteins to phosphorylated Y139 hypothetically provided new functions and potentially a new platform for eumetazoan evolutionary innovation. This system may have produced the gastrulation organiser of eumetazoans, which formed the foundations upon which subsequent chordate CpG epigenetic gene regulation evolved to regulate tissue-specific gene expression.
While the putative coiled-coil helical MIHIR region is not helical in the PGRMC1 crystal structure [4], the evolutionary conservation of Fig. 9B suggests evolutionary selective pressure to form a helix. If a helix does form, since the MIHIR is situated immediately between two helices which make direct heme contacts [4], it is highly likely that allosteric conformational changes throughout the protein would alter the geometry of heme-interacting residues and reduce affinity for heme. Indeed Y113 (the tyrosinate heme iron chelating residue [4]) is among the most frequently observed phosphorylation sites for PGRMC1 [66], and phosphorylation of Y113 would be incompatible with heme binding. Y113 is also the phosphate acceptor of a conserved YxxL/I immunoreceptor tyrosine-based activation motif (ITAM) [5, 220]. Phosphorylated ITAMs were first associated with membrane trafficking of non-catalytic tyrosine-phosphorylated receptors [221].
Accordingly, it is fully appropriate to think of PGRMC function in terms of
heme-dependent and -independent different roles for apo- and holo-PRGRMC. Any
association between the MIHIR, membrane trafficking, or motility remain
hypothetical and must yet be experimentally explored. However, the membrane
trafficking role of PGRMC1 is involved in the pathogenesis caused by synaptic
oligomers of A
Understanding the evolutionary origins of TMEM97/PGRMC1 interactions may be instructive in rationalising the system-wide effects of the system, such as roles in disease and pathology. Two regions of PGRMC1 seem to offer the most promising potential TMEM97 interaction sites.
The predicted negatively charged MIHIR alpha helix of Fig. 9C provides a potential interaction site between PGRMC1 and TMEM97. According to the AlphaFold in silico structural prediction [224, 225] and the crystal structure [226] (https://alphafold.ebi.ac.uk/entry/Q5BJF2), the TMEM97 protein contains four transmembrane helices, and a cytoplasmic C-terminal region in solution consisting of residues 162-176 with sequence YKYEEKRKKK. This polybasic sequence could quite conceivably interact electrostatically favourably with the putative MIHIR helix, to the extent that it even has five positive charges to match the five negatives of the MIHIR helix of Fig. 9C, and also C-terminally adjacent negative charges which could interact with the C-terminally adjacent positive charge at position 2 in the MIHIR helix of Fig. 9C. While attractive, this hypothesis requires experimental testing. It predicts that the PGRMC1 MIHIR region is induced into a coiled interaction with another protein that may bind some myosin motor proteins alternatively to PGRMC1, and that the resulting negatively charged exposed helical surface of the PGRMC1 MIHIR interacts with the C-terminus of TMEM97.
Given that the interaction between PGRMC1 and TMEM97 is associated with enhanced endocytosis of the low-density lipoprotein receptor (LDLR) [149, 150], it is tempting to extend the hypothesis to include the proposal that the unknown myosin-binding protein is associated in some way with recruitment of the actin-based motive force required for vesicle internalisation, and that therefore the MIHIR, a eukaryotic MAPR invention [8], is involved with PGRMC1’s membrane trafficking function. Furthermore, Y139 phosphorylation would be predicted to negatively regulate that function.
An alternative candidate region of PGRMC1 to interact with the C-terminal TMEM97 YKYEEKRKKK region is the PGRMC1 C-terminus. The PGRMC1 region from 180-195 (YSDEEEPKDESARKND) has suitable distribution of positively and negatively charged residues for electrostatic interaction. Precisely that region was absent from the PGRMC1 crystal structure [4], indicating a conformationally less restrained region. These alternative possibilities should be studied in the future.
Characterization of the TMEM97 interaction site should be quite informative about PGRMC1 biology. If TMEM97 interacts with the MIHIR, it suggests an ancient biology which predates animals, and could even trace back to the earliest eukaryotes (if the putative TMEM97 interaction does not require phosphorylated PGRMC1 Y139, which evolved in early animals [7]). On the other hand, a TMEM97 interaction with the PGRMC1 C-terminus would indicate biology associated with the evolution leading up to the earliest eumetazoan, since the C-terminus was absent in the common ancestor of earlier branching animal groups such as poriferans and ctenophores, and was only present in the common ancestor of placozoans (which do not have Y180) and eumetazoans (which have Y180) [7].
The LDLR system of lipid transport through the blood is obviously an invention of multi-tissued eumetazoans, such as chordates, with circulatory systems. However, a TMEM97/PGRMC1 mediated mode of vesicular endocytosis could have existed since early eukaryotic times.
In a very rudimentary introduction to the evolution of animal body plans (see Nielsen [212] for an authoritative account), the first multicellular organisms possessed epithelial layers surrounding a lumenal cavity, which arose by division of a single fertilized cells. The shape of the surface could be altered using the motive force of coordinated cytoplasmic actin skeletal forces applied across adherens junctions, which is how the body plan of placozoans and cnidarians are formed (Fig. 11C,D). The LEUMCA developed the first gastrulation organizer, which (via adherens-junction mediated actomyosin morphogenic force) invaginated a portion of the epithelia to form a cavity called the archenteron (Fig. 11, Ref. [227, 228, 229, 230, 231, 232]). This was associated with the appearance of a new pathway for the induced differentiation of new cell types that were absent from earlier animals, including nerves, muscle, and photoreceptor cells.
 Fig. 11.
Fig. 11.Eumetazoan gastrulation and body forms. (A) Development of the
blastula from the zygote. The right image shows a section through the blastula,
to reveal its hollow nature and interior blastocoel cavity. (B) Onset of
gastrulation near the organizer, which forms the archenteron cavity that will
develop into the adult gut. Wnt/
The LEUMCA has been historically considered as the first diploblastic organism, with a body derived from an ectodermis (that gave rise to nerves and epidermis), and an endodermis (that generated an adult with gut and associated gland tissues, as well as some nerves). Bilaterian animals have long been recognized to possess three germ layers: endoderm, ectoderm, and mesoderm. Of these, the genes expressed in bilaterian endoderm show greatest similarity to single celled metazoan sister group choanoflagellates. Ectoderm is proposed to have arisen from endoderm by loss of the ancestral feeding specialization, followed finally by the evolutionary appearance of mesoderm [195, 233, 234].
Based upon the expression of related transcription factors between cnidarian pharyngeal ectoderm and the endodermal midgut of bilaterians (including insulinogenic cells and the vertebrate pancreas), Steinmetz et al. [227] reported that bilaterian endoderm is related to cnidarian pharyngeal ectoderm, while cnidarian endoderm is related to bilaterian mesoderm [235]. This proposal has been followed in Fig. 11. Assuming that this model is correct, the LEUMCA already possessed discrete differentiated cell types with differential gene expression patterns that were the evolutionary precursors that produced the bilaterian three germ layers. However, these were distributed in a single invaginated (diplobastic) epithelial layer.
The concept of diploblastic cnidarians remains intact after this finding because there are only two cnidarian germ layers. However, the multiple different cell types induced after gastrulation by the LEUMCA appear to have already included a specialized endodermal cell type that would later give rise to bilaterian mesoderm. i.e., the proto-mesoderm was present, but manifest as just another region of specialized epithelial cell. In this model, the most conspicuous embryological innovation leading to bilaterian animals was the mesenchymal amoeboid transition (MAT)-induced mobilization of proto-mesodermal cells—related to the evolutionarily youngest cell-type of the LEUMCA (which forms endoderm in cnidarians)—to migrate into the blastocoel. That is, bilaterians undergo epithelial-mesenchymal/mesodermal transition (EMT) of their evolutionarily youngest early embryonic cell lineage. Such movement involves the response of the actin cytoskeleton and myosin II family motor proteins to locally induced or transduced stress, as well as specialized cell adhesion properties [236]. Critically, adherens junction-dependent epithelial cell contacts had to be disrupted for MAT to occur. Irrespective, we can assume that the gastrulation-induced differentiation of these novel cell types required organizer activity in the LEUMCA.
We are interested in the LEUMCA because it is the organism that acquired PGRMC Y139 and Y180, which were later strongly conserved in eumetazoans (Y139 appears to be slightly less conserved) [7]. Looking at the LEUMCA from a strictly PGRMC perspective, we might conclude that placozoan PGRMC does not possess Y139/Y180 (Fig. 8A,B), and hence the lineage leading to placozoans diverged before the LEUMCA evolved, and many multigene comparisons have supported this branching topology.
However, Laumer et al. [178] identified compositional heterogeneity of orthologous genes selected for analysis as a source of systematic error in such studies that attempted to reconstruct sequence-based phylogenetic trees. In attempting to compensate for this error, their revised analysis provided comparably strong support for placozoans belonging in the clade of LEUMCA descendants (as a sister group to the cnidarians) as support for placozoans branching earlier than cnidarians and bilaterians. Subsequent studies reinforced the potential existence of a placozoan/cnidarian clade [176, 179], however if we demand that the last eumetazoan and placozoan common ancestor (LEPCA, Fig. 8A) underwent gastrulation, for which there is no current evidence for any ancestor of placozoans, then the actual status remains unresolved. Examples of some markers supporting the monophyly of the placozoan, cnidarian and bilaterian clades include the transcription factor Hif-1 [53, 212] the Allatotropin/Orexin G protein-coupled receptor family [237], and certain co-inherited paralogous gene duplication products [176]. A C-terminal extension to PGRMC [7] is also common to these groups (Fig. 8B).
If placozoans are indeed descended from the LEUMCA then they must have followed an evolutionary path where the organizer function and advanced body grade (e.g., gut and nerves) of the LEUMCA were lost. With loss of the gut, this presumably would have involved reversion to primitive states, such as a mode of feeding reflecting the pre-LEUMCA state where nutrients are taken up across the epithelia [238]. If so, the concomitant loss of the PGRMC tyrosines would argue strongly for their requirement for organizer function and conventionally accepted eumetazoan identity.
In consideration of the body plan of the LEUMCA, we may also be considering the first bilaterally symmetrical organism, since there is debate over whether cnidarians were originally bilaterally symmetrical, with secondary loss by all cnidarian groups except anthozoans (sea anemones) [239]. Here then, we must unfortunately distinguish between the first bilaterally symmetrical animal (which may or may not have been the LEUMCA, possibly including the ancestor of placozoans), and the last common ancestor of bilaterians, being the urbilaterian organism that gave rise to the group currently referred to as Bilateria, consisting of Xenacoelomorpha (primitive triploblasts) and Nephrozoa (protostomes and deuterostomes, the latter of which includes humans) [179, 240].
Historical assumptions that central nerve cords of diverse bilaterian animals were inherited from a common ancestor have recently been questioned. Remarkably, nerve cords and body segmentation mechanisms appear to have evolved independently in different bilaterian lineages [241], however the case is still debated [242].
The common bilaterian ancestor appears to have had a system of mediolateral (middle to side) patterning. It was also endowed with a neurogenic ectoderm which may have given rise to dorsoventral polarity [242]. If we assume that bilaterian nerve cords evolved independently (parallel evolution), and that cnidarians were ancestrally bilaterally symmetrical, then the LEUMCA is expected to have been bilaterally symmetrical with a non-centralized nervous system, perhaps resembling that of cnidarian nerve nets, and a blind gut. If we do not accept independent nerve cord evolution then bilaterality probably arose later in a worm-like organism [239], and the LEUMCA would have been simpler.
Evans et al. [230] have recently described the so far earliest known bilaterian from the Ediacaran fauna. It was a benthic organism that burrowed in oxygenated shallow silt on the ocean bed. Fig. 11F is (perhaps incorrectly) reconstructed on this organism, assuming no urbilaterian central nerve cord and centralized anterior photoreceptors. LEUMCA photoreceptors develop using some genes conserved with bilaterian eye development [243]. The anterior end of the bilaterian organism described by Evans et al. [230] was consistently wider than posterior, suggesting some degree of neural cephalization.
The LEUMCA probably excreted toxic metabolites across its endodermal digestive cells [244]. It is accepted that the LEUMCA developed the gastrulation organizer (whether or not placozoans secondarily lost the organizer). This enabled the appearance of new cell types, which have been termed ‘apomeres’ (essentially analogous to apomorphies from paleontology, but at the cell-type level, and distinguished largely by identifiably inherited patterns of transcription factor activity), such as the neuronal apomere. The subsequent evolution of new animal apomeres not only permits us to reconstruct animal evolution by tracing the presence of similar cell types [245, 246], but also was enabled by the gastrulation organizer that appeared concurrently with PGRMC Y139 and Y180.
In considering the characteristics of the LEUMCA, it is only fitting to turn first to gastrulation, which refers to the embryological invagination of some cells from a topologically ball-like embryo (itself an animal innovation) to generate a cup-shaped intermediate with an outer ectoderm and inner gastroderm (Fig. 11A–C). All animal phyla undergo gastrulation, including sponges, however new cell types do not originate from the formation of the cup like indentation formed by gastrulation [196, 247].
For the sake of completion, note that the diagram depicted for the cnidarian in Fig. 11A–D is certainly an oversimplification. The jellyfish Aurelia appears to undergo a “secondary gastrulation” during metamorphosis from planula larva to polyp, during which larval endoderm is replaced by newly generated tissue [248]. Similar destruction and redevelopment during cnidarian metamorphosis has been observed for nervous system [249, 250], muscles [251] and tentacles [252, 253]. However, consideration of such cnidarian developmental complexity exceeds the purview of this work.
The LEUMCA was the first organism where a differentiated gastroderm persisted to adulthood and was associated with the induction of new patterns of gene expression in some cell types to generate new cell types [247], enabling for the first time a determinated body plan with an extended number of standardized morphological features achieved through a regulated process of cell differentiation. It may be useful to forget the complexity of bilaterian animals and recall that the LEUMCA organizer gave rise to a very primitive organism that would probably be nonviable today, but in its day was the pinnacle of multicellular sophistication. However, it already possessed many of the genes involved in germ layer specification upon which the later bilaterian evolutionary radiation would be based [235].
Importantly, the LEUMCA grade of organization provided an evolutionary springboard, described by Arnellos and Keijzer as “a qualitative jump—a major transition”, from a world of animal movement driven by cilia and contractile epithelia (adherens junction-mediated actin movements) to one of contraction-based (muscular) motility. The organizer was the enabling phenomenon for the evolution of the body forms we find from the very earliest animal fossils of the Ediacaran fauna (Fig. 8C), and later led to the evolutionary extravaganza known as the Cambrian explosion [74], which apparently was also fueled by a period of increased oxygen and biomatter availability compared to Ediacaran times [174].
That this scenario for PGRMC1 function is correct was supported by the recent
report of Lee et al. [254] that at ER-plasma membrane junctions PGRMC1
(and also PGRMC2) directly bound to a coiled-coil motif of stromal interaction
molecule 1 (STIM1), an EF-hand domain protein which acts as a Ca
We have previously demonstrated that PGRMC1 phosphorylation mutants affect cell migration [50], and we proposed that the PGRMC1 MIHIR contained a coiled-coiled motif [7] which should be regulatable via Y139 phosphorylation since Y139 constitutes one of the hydrophobic coiled-coil heptad repeat residues (Fig. 10B). Taken together, these results suggest that PGRMC-dependent and regulatable focal adhesion activity via adherens junctions and associated regulated cell migration appeared in the LEUMCA along with and dependent upon a new structure of the PGRMC gene.
Having considered LEUMCA gastrulation, it is imperative that we understand the LEUMCA and its organizer, from which our own branch of the evolutionary tree developed. Descendants of the LEUMCA are called eumetazoans because of their adult possession of a gut and differentiated cell types, including the first neurons (excluding the independent ctenophore development of neuron-like cells) [192]. The group is also sometimes referred to as Gastraea or Gastraeozoa (“animals with an intestine”).
Cnidarian organizer function is not restricted to embryogenesis of a zygote. For instance, in Hydra, pluripotent stem cells are scattered throughout the body. These can permit parthenogenetic formation of new offspring. Even more impressively, a Hydra polyp can also be bisected into multiple fragments (e.g., 20), each of which will grow into a complete adult with all specialized cell types [216]. Therefore, organizer function can arise from multiple stem cells within the Hydra body.
Although the mechanics of gastrulation differ markedly among cnidarian groups
[255], it is clear that eumetazoan animals inherited a gastrula life stage from
the LEUMCA, and its invention of the organizer [212, 231, 235, 239, 256]. The
blastula region on the opposite side to the point of sperm entry becomes the
organizer (Cell divisions initiated by formation of the zygote nucleus induce its
activity there). The organizer induces epithelial cells to undergo
differentiation in a process involving bone morphogenetic protein (BMP) factor
and Wnt/
Brachyury in the LEUMCA, as interpolated from the cnidarian state, activated
ectodermal genes, and repressed endodermal genes, and this later evolved into the
bilaterian system where triploblasts develop mesoderm from Brachyury-expressing
ectoderm [263, 264]. In the early mouse embryo, a structure called the primitive
streak forms at mouse embryonic day 6.5, at the time that primed PSCs appear
before implantation. Its cells undergo the Wnt/
PGRMC1 and PGRMC2 are expressed in vertebrate oocytes [14, 26, 57, 60, 145, 182, 267, 268, 269, 270, 271, 272, 273, 274] as well as spermatozoa [23, 275], where PGRMC is involved (directly and/or indirectly) with P4 responsiveness (Indeed, this has been one of the most fertile areas of PGRMC1 research, excuse the pun). Importantly, both PGRMC1 and PGRMC2 are expressed from the zygote to blastula stage in mammals [276, 277], and so are present at the onset of gastrulation. In the nematode Ceanorhabbitis elegans the PGRMC homologue Ventral Midline-1 (Vem1) as well as the MAPR NEUFC family member Vem2 are also expressed from the oocyte stage and fertilized zygote, through all developing cell stages to the blastula [234] (Also see Wormbase [278] entries for both proteins: (WBGene00006478#0-9g6e1bcd-10, and WBGene00006890#0-9g6e1bcd-10). We may conclude that the PGRMC gene in the LEUMCA was similarly expressed from zygote to blastula, and therefore was available to participate in organizer determination and activity.
Brachyury is one of the genes that arose during single celled holozoan evolution, which deserves special attention here in the context of metazoan gastrulation and the protozoan phenotypic switching between flagellated and ameboid morphotypes. The filasterean holozoan single-celled organism Capsaspora owczarzaki exhibits dynamically interconvertible histone-modified chromatin states that regulate transcription factor cis-regulatory elements of gene promoters. These are associated with altered gene expression of coregulated genes that characterize different stages of the life cycle. The Brachyury transcription factor is encoded by one of the genes acquired by the common ancestor of Capsaspora and animals. It is a member of the T-box transcription factor family. A T-box protein was present in the ancestor of opisthokonts (the phylogenetic group that contains fungi and animals), but the Brachyury type of T-box protein was first acquired by the ancestor of filastereans, such as Capsaspora [279], and animals.
Capsaspora has a life cycle consisting of three cell morphotypes, one of which is amoeboid and motile [280]. Brachyury induces a program of enhanced actin-dependent amoeboid migration in Capsaspora that involves similar genes used by amoeboid mammalian cell motility [281]. These functions are essential in animal gastrulation and EMT-induced mesoderm formation [282], and the Capsaspora Brachyury protein can at least partially induce gastrulation in Xenopus [279]. Brachyury expression in cancers induces EMT, which is strongly associated with malignancy [283]. It appears that the LEUMCA harnessed suites of genes that had been present for hundreds of millions of years. By the innovation of gastrulation, gene activity was controlled by a novel mechanism of genomic packaging. It is possible that PGRMC1 tyrosine phosphorylation played a foundational role in that packaging mechanism, which deserves to be investigated in the future.
Expression of PGRMC1 in MES-SA uterine sarcoma cells led to EMT [16]. A potential PGRMC1 connection with the Brachyury and its actin cytoskeleton regulation is reinforced by the discovery that the MIHIR of vertebrate PGRMC1 contains a predicted coiled-coil motif related to one found in the coiled-coil region of multiple myosins [7].
As discussed above, Y139 phosphorylation may affect PGRMC1 interactions with the actin cytoskeleton (experimentally unverified). Furthermore, in human cell culture the mutation of presumed negative regulatory S57 and S181 residues (both adjacent to Y180 in the folded protein structure [66]) caused increased cell migration in scratch motility assays, and elevated expression of a set of proteins associated with actin cytoskeleton. This was associated with activation of the PI3K/Akt pathway, and efficient subcutaneous mouse xenograft tumor formation. Upon additional mutation of Y180 the elevated actin cytoskeletal proteins, the enhanced motility, the PI3K activation, and efficient xenograft tumor formation were all absent [50]. The effects appear to have been mediated by altered PGRMC1-dependent genomic epigenetic gene packaging [19].
Since 2012 we have known that PGRMC1 could affect gene transcription, since its depletion resulted in elevated expression of a large number of genes [143]. It is likely that epigenetics was involved, however Sumoylated PGRMC1 directly binds and activates promoters with binding sites for the transcription factor T-cell factor/lymphoid enhancer-binding factor (TCF/LEF), a target of the Wnt pathway [142, 143, 144]. Taken together, these results suggest that PGRMC Y180 phosphorylation could regulate or is involved in Brachyury function during organizer function, which merits further investigation.
We have also recently demonstrated that PGRMC1 is found in immune precipitation complexes with many proteins associated with the actin cytoskeleton, and that the small molecule inhibitor AG-205 prevents many of those interactions [91]. Salsano et al. [172] also identified many proteins associated with actin cytoskeleton and membrane trafficking to be co-precipitated with PGRMC1. However, it remains to be demonstrated that actin interactions are via the MIHIR region. Saliently, PGRMC1 affects both actin cytoskeletal protein abundances and cell motility or migration [50], many instances of which are associated with EMT and/or tumor metastasis [16, 50, 284, 285, 286, 287, 288, 289].
A working hypothesis emerges where PGRMC-dependent alterations in actin cytoskeletal regulation during EMT modulate adherens junction-mediated epithelial cell adhesion with accompanying enhanced cell motility, enabling the epithelial to mesenchymal transition that underpins bilaterian evolution, and the development of mesoderm. In other words, the acquisition of PGRMC1 Y139 and Y180 by the LEUMCA may have facilitated gastrulation, and the evolution of eumetazoans.
However conjectural this may be, the hypothesis predicts that PGRMC1 may
regulate proteins of the adherens junction. PGRMC1 phosphorylation status can
modulate vinculin abundance [50], and the small molecule AG-205 disrupts protein
complexes involving PGRMC1 and proteins of the actin cytoskeleton (which
contained calponin homology (CH) actin-binding domains), among which
The
Conceptually most important here is that the combination of PGRMC Y139/Y180 tyrosine phosphorylation and the functionality of the extended C-terminus must be essential for some feature developed newly by the LEUMCA, which was inherited by bilaterians, because those residues are conserved in eumetazoans. Since the LEUMCA did not possess complex organs or vasculature, the number of potential roles is hopefully of a manageable order of complexity to consider with some modicum of systematics. Potential roles (in common with bilaterians) broadly include gastrulation itself, a gut, glandular (e.g., insulinogenic) cells, photoreceptors, muscle, and their organismal coordination by nerves (as well as their presumed maintenance during sleep). What seems most obviously important will be discussed in the following. This list may, of course, be conceptually incomplete.
The LEUMCA was the first animal where the embryological gastrula stage persisted to adulthood as a specialized gut, having spawned a suite of different specialized cell types in the process [247]. The gastro-epithelial surface is thought to have developed muco-ciliary secretions and to have been folded into gastric pouches which optimized nutrient absorption and permitted extracellular digestion [294]. This created a novel environment for bacterial colonization. Bacteria subsequently played a crucial part in the evolution of animals, with specialized microbial communities colonizing ectoderm and gut endoderm of cnidarians (reviewed by [295]) and triploblastic bilaterians. In the latter the gut forms a microbe-filled tube where bacteria ferment digested food into products that can be used by the host animal under anoxic conditions from worms [296] to mammals [297]. That coevolutionary journey between bacteria of the microbiome and host animals was born with the LEUMCA and its development of the specialized gut.
In mammals, colonic epithelial cells (colonocytes) consume oxygen by oxidative phosphorylation to promote gut lumen hypoxia associated with obligate anaerobic healthy gut microbiota. Failure to maintain hypoxic levels permits the expansion of facultative anaerobes, leading to dysbiosis that is associated with several pathologies [297]. In this process, the progenitor crypt stem cells perform Warburg glycolytic metabolism. Recall that PGRMC1 directs Warburg glycolytic metabolism [49, 50]. The differentiation process of mammalian gut epithelium involves a metabolic switch to activation of fatty acid catabolism and oxidative phosphorylation by mature colonocytes. Therefore, glucose metabolism is crucial in healthy gut function [297], and this dichotomous gut cell metabolism may be inherited from the LEUMCA. This sounds like a system that (hypothetically) could have been driven by PGRMC since LEUMCA times. We will consider glandular cells of the gut in a separate section.
Innervated striated muscles are present in both cnidarians and bilaterians, however these have different architectural organization and have been shown to have evolved independently in both lineages from a pre-existing suite of epitheliomuscular LEUMCA contractile apparatus [298]. We must conclude that the LEUMCA is not likely to have possessed striated muscle. Cnidarian muscle is innervated by neurons belonging to a nerve net with sensory neurons that enable rapid detection of and response to environmental stimuli that is superior to those of earlier-branching animals [74] (We will deal with neurons below).
However, the biology of the LEUMCA and its descendants is intimately associated with contraction-based motility that enabled predation of food that utilized the new gastric cavity [74]. The LEUMCA was probably benthic (sea floor-dwelling) and used cilia and mucous to trap organic material [247, 294]. It is then perhaps best to imagine coordinated organism-wide cellular epitheliomuscular contractions not yet at the level of striated muscle but providing sufficient locomotion to successfully forage for food. Cnidarians generate peristaltic waves via radial muscle contractions based upon coordinated adherens junction contractions [299]. The earliest known identifiably bilaterian fossil is thought to have burrowed through shallow nutrient-rich ocean sediments using peristaltic motion [230], which would have required a system of longitudinal and transverse muscle-like fibers.
Insulin-related peptides are conserved in deuterostomian, ecdysozoan, and lophotrochozoan bilaterian species, as well as cnidarians [300]. Therefore, they can be extrapolated back to the LEUMCA. The reconstructed LEUMCA possessed insulinergic glandular cells expressing the forkhead box transcription factor A (FoxA), which gave rise to related cnidarian (Fig. 12B, Ref. [216]) and mammalian (pancreatic) gland cells [227, 228, 229]. Insulin controls glucose homeostasis. PGRMC1 is known to regulate plasma membrane translocation of the insulin receptor, glucose transporters GLUT1 and GLUT4 [301], as well as glucagon-like peptide-1 (GLP1) receptor (GLP1R) [302], all of which are associated with hormonal glucose homeostasis. 3T3L1 pre-adipocyte stem cells also upregulate glucose intake by PGRMC1-dependent translocation of GLUT4 to the plasma membrane, where PGRMC1 and GLUT4 are co-localized and can be co-immunoprecipitated [162].
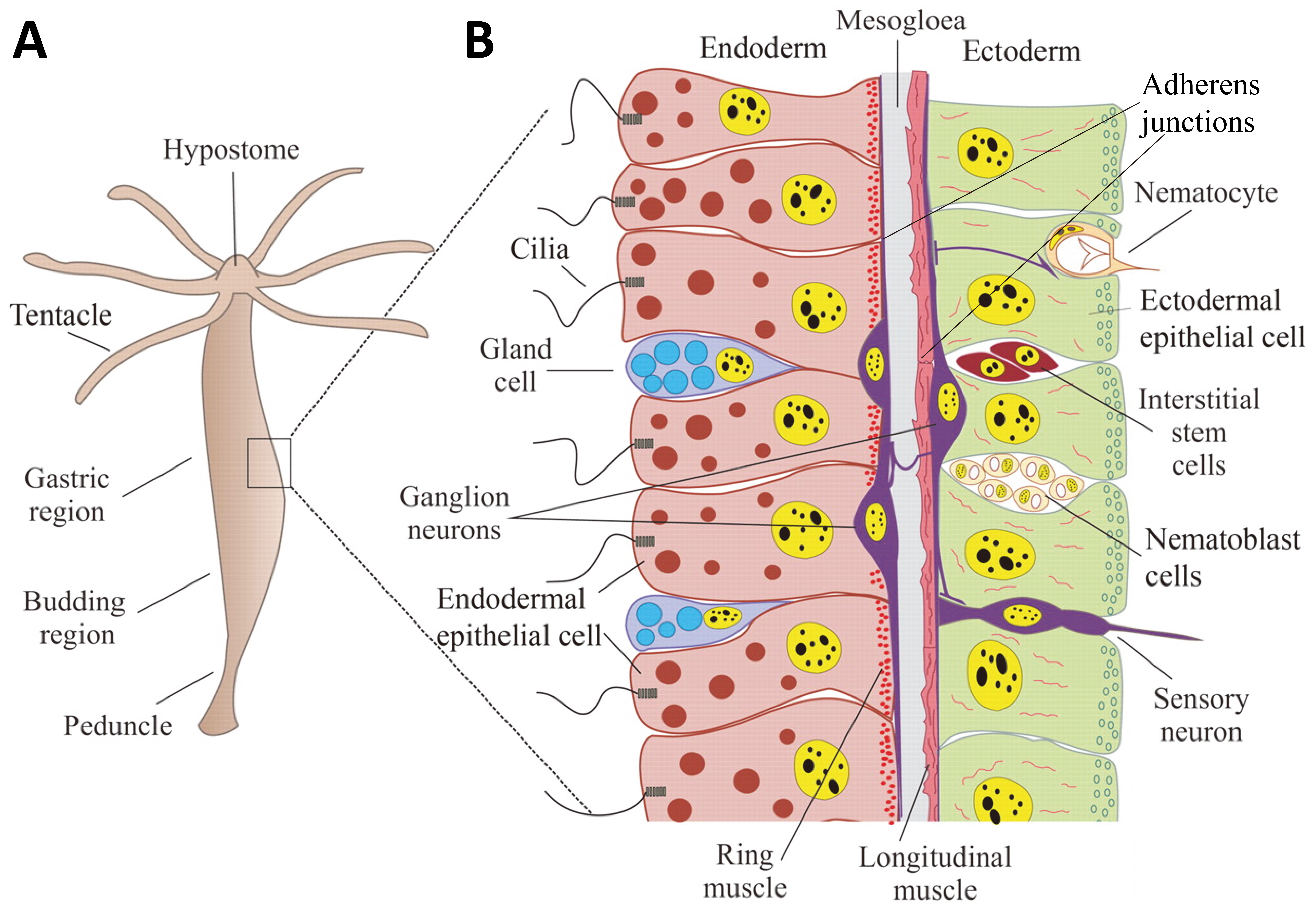 Fig. 12.
Fig. 12.Anatomy of a cnidarian hydrozoan polyp. (A) A Hydra polyp is essentially a two-layered tube, with a ring of tentacles around the mouth opening at the tip of the hypostome. Asexual budding occurs on the lower half of the body column. Interstitial stem cells and nematoblasts are distributed evenly in the body column, below the tentacle ring and above the border of the peduncle, which is the stalk between the budding region and pedal disc. (B) The bilayered cellular organization of a Hydra polyp. Ectoderm and endoderm are separated by an acellular matrix called the mesoglea (gray). All epithelial cells in Hydra are myoepithelial, with myofibers on the basal side (red). In ectodermal epithelial cells (green), the fibers are oriented longitudinally, and in endodermal epithelial cells (pink) they are oriented circumferentially (ring muscle). Most interstitial cells and nematoblast clusters are located between ectodermal epithelial cells. Neurons are found in both the endoderm and ectoderm. Sensory neurons are located between epithelial cells and connect to ganglion neurons (purple), which are at the base of the epithelium on top of the myofibers and sometimes cross the mesoglea. Different types of gland cells, most of which are found in the endoderm, are intermingled between the epithelial cells. Modified figure and directly copied legend from Technau and Steele [216], with permission of the publisher (Company of Biologists Limited, Cambridge, England).
Mammalian GLP1 is a peptide derived from the pro-glucagon precursor. This precursor protein encodes several different and in part overlapping incretin proteins which are generated by differential proteolytic cleavage, including glicentin, glicentin-related polypetide, oxyntomodulin, glucagon, GLP1 and GLP2 [303] (And see UniProt P01275). While glucagon is produced by pancreatic A cells under low blood glucose conditions, GLP1 is produced post-prandially by intestinal enteroendocrine L cells, pancreatic A cells, and the central nervous system. A hypothesis can be proposed here that this system represents an evolutionary descendant of PGRMC functions from the LEUMCA that mediated communication between cell types similar in complexity and spatial organization to those of Fig. 12B.
Oral glucose administration evokes a much stronger insulin release, due to intestinal GLP1 release, than is evoked by intravenous glucose injection. GLP1 acts on the pancreas to induce insulin secretion and on the central nervous system leading to appetite reduction (for reviews: [303, 304, 305]). As such, PGRMC is positioned to regulate energy communication between cells of bilaterian animals, which is possibly inherited from LEUMCA glandular cells.
This is a hypothetical proposal, so far undemonstrated. Future research should address the evolutionary history of the association of PGRMC and its Y139/Y180 phosphorylation residues with this system. However, it is notable that PGRMC1 also regulates glucose metabolism on a cellular level in response to P4, seemingly independently of glucagon-insulin signaling (although this has not been formally examined) [49, 306] and glycolytic metabolism is affected by PGRMC1 phosphorylation [50].
One relationship between glucose metabolism and the new LEUMCA requirement for
synaptic function could be due to membrane fluidity. Here, we need to briefly
introduce the fact that a class of serpentine seven membrane-spanning G
protein-coupled receptors are associated with membrane P4 receptor activity. A
complex involving PGRMC1, PGRMC2 and PAQR7 (or mPR
PGRMC1 also regulates the cell surface availability of a variety of other known proteins [47], including the insulin receptor [301]. What has this to do with membrane fluidity? PAQR-2 and insulin growth factor receptor-like 2 (IGLR-2) modulate membrane fluidity in response to glucose levels by regulating fatty acid desaturation levels in nematodes [308] and mammals [309]. Membrane fluidity is important for synaptic vesicle trafficking, as well as for the cell surface localization of receptors involved in ligand-dependent guidance of cell movement. PGRMC1 is implicated in fatty acid synthesis [41, 162, 163], and both PGRMC1 and PGRMC2 affect the activity of fatty acid 2-hydroxylase that is required for the synthesis of 2-hydroxylated sphingolipids in the nervous system and other cell types, which has been proposed to reflect PGRMC-dependent functional interdependence of sphingolipid and sterol biology on membrane properties [164]. It will be interesting to see whether PGRMC1 is involved in this possible modulation of membrane fluidity in response to insulin/glucagon hormonal regulation that could have been inherited from the LEUMCA.
Insulin affects synaptic function and the plasticity required for long-term potentiation [310], and pronounced hippocampal glucose consumption has long been known to be associated with learning and memory formation [311]. Accordingly, impaired glucose metabolism is associated with several neurodegenerative conditions [312, 313]. Therefore, regulation of glucose metabolism by PGRMC could have been associated with the evolution of the nervous system and ligand-guided cell migration. Having now been diverted from endocrine regulation of metabolism towards neural function, it is appropriate to consider neurons in more detail.
How the first synapse evolved has been unclear because most of the genes involved in synaptic function predated metazoans and are present in poriferans and placozoans that lack nervous systems [208, 245]. As an aside, this could be an indication of the importance of PGRMC tyrosine phosphorylation in the LEUMCA for the origin of the nervous system (Note, PGRMC is present in poriferans but was not classified as being among the group of genes analyzed as a ‘synaptome’ [208], and so was in any case not under the radar of these studies).
Be that as it may, based upon the expression of conserved common sets of transcription factors, the gastrulation stage embryonic LEUMCA also already possessed two regions of neurogenic tissue at opposite poles that are recognizable in its modern cnidarian and bilaterian descendants. These are proposed by Arendt et al. [247] to have given rise to an apical nervous system, involved in sensory function, and a blastoporal nervous system, involved in sensory neuron, interneuron and motor neuron formation, and exercising control over gastric function. They propose that derivatives of these two nerve centers from the LEUMCA became fused to generate bilaterian central nervous systems [247]. We have seen above that bilaterian nerve cords probably evolved convergently in different bilaterian lineages [241]. Nevertheless, Arendt et al. [247] equate the hypothetical LEUMCA dual-centered neurogenic nervous system with the central nervous system of mammals, which fused at the forebrain.
Consistent with a role in neurogenesis, we have long known that PGRMC plays an essential role in axon guidance during embryonic migration of central nerve cord neurons from nematodes to mammals, where PGRMC interacts with a member of the DCC family [314, 315]. This is perhaps related to the membrane trafficking function of PGRMC1 leading to cell surface localization of DCC to enable DCC to interact with extracellular Netrin guidance ligands, although this has not been demonstrated.
P4 leads to axonal outgrowth and improved learning in the adult mammalian brain, and PGRMC1 is associated with induced growth cones of nascent axons, which mediate the axonal navigation required for proper synaptic connections [316]. PGRMC1 is expressed widely in the cerebellum, especially during the neonatal period when synaptogenesis is enhanced, where it (under its 25-Dx synonym) is thought to be involved in dendritic growth, spinogenesis, synaptogenesis, and neurosteroidogenesis in Purkinje cells (a steroidogenic neuron type) [24]. So PGRMC1 directs axon migration leading to synaptic connection from embryology to adulthood. It is also present in neurite projections of adult neurons [317], is involved in synaptic function [63, 64], it can neuroprotectively regulate inflammation and neuroimmune function in a P4-independent manner [318], and PGRMC1 is tyrosine-phosphorylated only in post-synaptic density fractions of mouse brain [319].
These are precisely the suite of characteristics we could expect if PGRMC was a protein that originally enabled neurogenesis in the LEUMCA and is now so foundational to neurobiology that its functions have become indispensable for eumetazoans. Glial expression of PGRMC1 is required for neurite outgrowth [288, 320], a process that is essential to synapse formation and the establishment of neuronal circuits required for learning [321]. PGRMC1 is involved in the P4-dependent reorganization of the actin cytoskeleton that is critical to these processes [322].
Perhaps most poignantly for synaptic origins, PGRMC1 membrane trafficking is
required for the mechanism of action of new anti-AD drugs that displace
A
Phylogenetic analyses provide some support for the involvement of PGRMC tyrosines with Netrin/DCC involvement in neurogenesis. PGRMC phosphorylated tyrosines 139 and 180 [7] as well as both Netrin and DCC all arose in the LEUMCA, at the same time as neurons [54].
The other MAPR proteins Neuferricin and Neudesin [21, 327] also play roles in axon migration, which logically ends with synaptogenesis. Neuferricin [328] and Neudesin [329] are not only neurotropic, but also both promote neurogenesis. All three MAPR families obtained extended C-termini in the transition from pre-metazoan to LEUMCA, and PGRMC gained its Y139 and Y181 phosphorylated tyrosines in the LEUMCA [7]. Effectively, the post-LEUMCA PGRMC gene was a new gene with old and new functions and is an excellent candidate to explain the ability of nerves to form synapses. It is highly likely that PGRMC is involved in synaptogenesis, although this requires formal demonstration. However, all three MAPR families were possibly involved in the origin of nerves and synapses.
In mammalian synapses the post-synaptic membrane is of the order of 40% cholesterol, as are synaptic vesicles [330]. The human brain accounts for 2% of human body weight, but contains 25% of the body’s cholesterol [331]. Cholesterol is important for lipid rafts that enable receptors to function correctly, synapse formation and the propagations of action potential, and perturbed cholesterol metabolism has been associated with several neurodevelopmental and neurodegenerative diseases [331, 332, 333].
Because of the central role of PGRMC1 in cholesterol biology, it almost certainly dramatically affects membrane fluidity and synapse function by regulating cholesterol levels and/or changing fatty acid saturation. This can affect the activity of membrane proteins such as neurotransmitter receptors by allosteric interactions with cholesterol, as well as change the thermophysical properties of the membrane itself [334, 335, 336]. These are precisely the types of roles that may have maintained the hypothesized intimate association between PGRMC1 and neural function since neurons appeared in the LEUMCA.
The origins of bilaterians were probably associated with restriction of neural progenitors to the ectoderm, however consideration of bilaterian neurogenesis exceeds the scope of this work and has been recently reviewed elsewhere [211, 337]. See Heger et al. [54] for a recent phylogenetic treatment of this field, including bilaterian origins of the neurotropic nerve guidance system. Suffice to paraphrase that work here to say that the last common bilaterian ancestor (or at least the deuterostome/protostome common ancestor, excluding Xenacoelomorpha) possessed a circulatory system, serpentine GPCR monoamine neurotransmitter receptors, and a complex brain center, however structures in modern deuterostomian, ecdysozoan and spiralian brains may not share homology (may have arisen separately from a more primitive ancestral state) [54, 211].
The Xenocoelomorpha, who form a sister group to deuterostome/protostome bilaterians, probably ancestrally had a diffuse nervous net, reminiscent of the ancestral cnidarian grade [211, 338]. Therefore, if we postulate roles for the involvement of Netrin/DCC and PGRMC tyrosine phosphorylation in both the evolutionary origins of LEUMCA neurons and embryological neurogenesis, we should invoke a function that predated the appearance of a central nervous system.
Mammalian neurons share functional interrelationships with associated glial cells such as astrocytes, and myelin-forming oligodendrocytes of the central nervous system or Schwann cell of the peripheral nervous system. Neurons rely on elevated rates of glycolysis upon activation, which appears related to rapid ATP requirements that outstrip available oxygen availability [339, 340].
Alzheimer’s disease is associated with astrocytes that provide metabolic and physical support for neurons including providing cholesterol and modulating synaptogenesis and synaptic function, and also regulate the myelination that is important in rapid long-range propagation of action potentials [341]. A similar system of neuron glial-dependent myelination and metabolic exchange is observed in Drosophila neurons [342, 343]. However, since mammalian myelin first appeared in jawed fishes [344], accompanied by the evolution of voltage-gated sodium channels with new properties [345], and since central nervous systems and segmentation appear to have evolved separately in different bilaterian lineages [241], it is probable that insect myelinated nerves represent parallel evolutionary acquisition of similar cell states required for complex body organization, that were derived from the same toolbox of genes provided by the LEUMCA, but which are most probably not the same cell type apomeres. That is, mammalian and insect glial myelin cell states are probably examples of convergent evolution, and not induced by related gene expression patterns controlled by coordinated transcription factor networks that are inherited by common descent, as defined by Arendt et al. [245]. However, a recent study identifies PGRMC1 as one of a suite of genes involved in neurosteroidogenesis, and metabolism and mode of action of P4, which are functionally implicated (increased expression) in post-natal myelination in the mouse cerebellum [346].
The LEUMCA, as judged by the presence of Hif-1 [53] and the metabolism of extant cnidarians [347, 348], was able to induce glycolysis and produce lactate in response to hypoxia. I am unaware of any study relating cnidarian neuronal function to glycolysis. However, the thesis being proposed here is that metabolic modulations associated with PGRMC may have been crucial in enabling nerves and synapses to evolve/differentiate in the eumetazoan clade of LEUMCA descendants which inherited the PGRMC Y139/Y180 configuration at a time of presumed changing oxygen concentration (Fig. 8A). As such, they may have been foundational to the evolution of PSCs. These themes are revisited in the accompanying paper in consideration of Alzheimer’s disease and aging [2].
In his book “A Thousand Brains: A New Theory of Intelligence” [348], and in several recent publications (e.g., [349, 350, 351, 352]), Jeff Hawkins and colleagues propose a revolutionary new model of cognitive intelligence where columnar units of tens of thousands of neurons can process environmental information to form multidimensional grid-like reference frame models of reality. These collectively contribute via a voting-like function to determine the model of conscious ‘reality’.
The human cerebral cortex has about 150,000 of these columns, each forming reference frames for different aspects of input (auditory, visual, olfactory, tactile, etc.), whose combined action contributes to consciousness, memory, and intelligent thought. Importantly for PGRMC proteins, the formation of dendritic synapses establishes the circuitry that potentiates the ability of each column to create reference frames for different modelled features. Most information of the nervous system is stored in the connection of neurons to other cells via synapses (some must be stored in the physical three-dimensional structure of axons and dendrites themselves). Such stored features of reference frames may range from sound to position in three-dimensional space, or even to higher conceptual constructions involved in mathematics or language in humans, with different columns and brain regions specializing in different features in humans.
The “thousand brains theory” posits that mammalian brains developed the neocortex by increasing (‘evolutionary cut and paste’) the number of pre-existing reference frame-generating systems such as those of the hippocampus of the pre-mammalian reptilian brain to create the neocortex, which is specific to mammals. These were then multiplied during evolution to increase intelligence and brain size in e.g., the primate lineage leading to humans [353].
Important for our consideration here is the presumption of this model that a pre-existing ancestral neural system could generate a reference frame which included a model of the organism, its state in three-dimensional space through time, and its relationship to its environment. Importantly, it needed to learn environmental variables such as proximity to food sources. Once this system existed, that ancestral reference frame-forming neural architecture provided the building block for complex central nervous systems.
Could the cnidarian neural net, as a surrogate model of the ancestral LEUMCA nervous system, already have served that function of forming a “one brain” reference frame of the state of the organism in its environment? Cnidarians are capable of cognition. They sense their environment, can be habituated/dishabituated, sensitized, and conditioned, and associatively learn about things [354]. These tasks involve receiving and integrating signals from sensory systems such as photo-receptors and epithelia to metabolic monitoring and coordinating responses such as muscular contraction and whether or not to release nematocyst stinging cells, which are expensive to build and must be replaced after one use.
It is reasonable to propose that the cnidarian nerve net can create a relatively simple yet sufficient multidimensional grid-like reference framework model of itself in its environment. Extrapolating back, the LEUMCA would then have already possessed the essence of the neural cognitive building block that was refined later to produce our cerebral neocortex. If so, the acquisition of PGRMC Y139 and Y180 of the LEUMCA may be related to the acquisition by eumetazoan animals of cognitive intelligence. Similarly, the perturbation of PGRMC function would lead to impaired cognitive ability [2].
This hypothesis makes predictions that to the author’s knowledge have not been examined. If it is correct, then existing double knockout mouse and fish models [55, 162, 163, 273, 274, 355, 356, 357, 358, 359, 360] are predicted to possess decreased synaptic number and impaired learning in addition to decreased fertility and perturbed lipid metabolism reported. In that case, the above-invoked neural function that predated the appearance of a central nervous system would be the formation of synapses that are associated with memory formation. Furthermore, we would expect changes in PGRMC tyrosine phosphorylation during the consolidation of neural pathways that we call memory. This happens in sleep.
Sleep is critical for neural function and memory formation [361, 362]. During sleep some synapses are weakened and others strengthened to consolidate recent memories in the form of neural excitation pathways [363].
Of relevance to our theme, synaptic changes occur during sleep, and sleep evolved at the same time as the organizer, i.e., in the LEUMCA. Cnidarians change behavior at night, which is considered as sleep because animals exhibit slower reaction times in this state, and if deprived they will later ‘sleep’ longer to catch up: a behavior known as homeostatic sleep regulation. This implies that sleep is related to a circadian-associated neural regeneration process that appeared before highly centralized nervous systems [364, 365, 366]. For the moment, note that sleep appeared evolutionarily at the same time as the gastrulation organizer, Netrin and DCC, the first neurons, and the combination of PGRMC Y139 and Y180.
Also recall that PGRMC2 knockout led to reduced heme shuttling to the nucleus,
which led to increased stability of heme-responsive transcriptional repressor
Rev-Erb
As a working hypothesis, we could do worse than propose that the coincident appearance of gastrulation, PGRMC tyrosine phosphorylation at Y139 and Y180, muscles, synapsed nerves, and photoreceptors, together with Hif-1, the Netrin/DCC (and Netrin/Neogenin) system and ‘sleep’ (diurnal behavioral changes) were all part of the emergent biology of the LEUMCA, dependent in good part on PGRMC tyrosines. If the LEUMCA migrated to the bright upper water layers by day to ingest the plentiful photosynthetic organisms there, they would have been localized by photodetection, hunted by neuronally-directed muscle motor propulsion, and transferred to a new gut. There, specialized epithelial cells enhanced digestion and absorption, probably assisted by the very first specialized gut microbiome, as the organism ‘slept’ to regenerate. It may also have been benthic, sifting through seabed detritus. This organism may have been primitive by modern standards, but its attributes would transform life on earth.
Conventional thought has attributed sleep regulation in insects to monoamine and peptide neurotransmitters [368], however it has long been known that the insect steroid ecdysone regulates sleep in the fruit fly Drosophila melanogaster [369]. It will be interesting to see whether Drosophila PGRMC (called membrane steroid binding protein, MSBP: UniProt Q9VXM4) regulates ecdysone production and sleep. The production of several reproductive hormones is affected by sleep and circadian rhythm in humans, which influences fertility (another area of PGRMC ‘core biology’). Likewise, reduced steroid levels lead to poor quality of sleep [370]. We will return to sleep and memory formation in consideration of gut flora below, and Alzheimer’s disease and circadian NAD (nicotinamide adenine dinucleotide) metabolism in the accompanying paper [2].
In the transition from earlier-branching animals such as ctenophores and poriferans to the LEUMCA, the PGRMC gene gained a C-terminal extension which contains the Y180 motif (Fig. 8B). Tsai and colleagues [371] from the University of Michigan Medical School have recently demonstrated that this region participates in a newly described function for PGRMC1: binding to low molecular weight complexes of misfolded proteins in the lumen of the ER and targeting them for ER-phagy (literally ‘ER eating’) via the reticulon-3 (RTN3)-driven targeting complex in HEK293 cells, Cos cells, and ex vivo primary pancreatic islets. In ER-phagy, portions of the ER are portioned off and sent to the endolysosome for degradation (for review: [372]). Notably, in this function PGRMC1 has the type 2 membrane topology observed in embryonic stem cells [373], Alzheimer’s neuron synapses [64], hepatocytes [374], and cultured lung cancer cells [375]. That is, the C-terminus is in the ER lumen, and hence unavailable to cytoplasmic enzymes such as kinases, ubiquitin conjugating enzymes, and the majority of CYP450 molecules that have cytoplasmic active sites [376]. The PhosphositePlus site contains hundreds of references of PGRMC1 with post-translational modifications of the C-terminus consistent with a type 1 orientation (https://www.phosphosite.org/proteinAction.action?id=5744). It remains unclear why PGRMC1 is found with different membrane topologies in different circumstances. The answer will probably illuminate a deep truth about eukaryotic biology.
The newly detected ER-phagy targeting role of PGRMC1 [371] adds another function to the dizzying functional list already attributed to this protein [47]. Our goal is to part the fog to discern an overarching biological model—a PGRMC1 raison d’être—that potentially explains the observations. The following is accordingly speculative.
ER-phagy is one path of disposal for misfolded ER proteins that is the end point of the unfolded protein response (UPR), one of the components of ER stress. In eukaryotic microorganisms (primarily concerning yeasts), ER stress involves the induction of ER-associated degradation (ERAD), the ER overload response (EOR), the UPR, sterol-regulated cascade reaction, autophagy, hypoxia signaling, and mitochondrial biogenesis (for review: [377]). ER-phagy operates alongside and communicates with those processes [372]. We have seen above that PGRMC1 has already been implicated in autophagy via complexes with MAP1LC3 and UV radiation resistance-associated gene protein (UVRAG) [138], and so we are conversant with this type of function. Autophagy is regulated by modulating membrane contact sites between different organelles [378], and PGRMC1 is required to maintain mitochondrial-ER contact in some cases [113]. The eukaryogenic endosymbiosis required the establishment of such membrane contact sites, and mitochondrially-synthesized membrane lipids are transferred via such contact sites to the rest of the cell. This is probably responsible for the closer resemblance of eukaryotic membrane lipids to bacteria than to the archaeal host cell membrane [379].
We also discussed above the relationship between hypoxia and sterol biology, how sterols are essentially eukaryotic bacterial hopanoids, and how PGRMC1 with Insig/SCAP regulates mevalonate pathway activity, while PGRMC1/CYP51A1 make the very first modification of a sterol. Here we note that sterol synthesis is a part of the ER stress response that is coordinated with mitochondrial biogenesis. Note that PGRMC1 is observed as an ER protein in many contexts, and so an overarching role in these processes is feasible [46, 374, 380].
The above responses apply primarily to eukaryotic microorganisms [377]. We can associate PGRMC proteins at several steps, as well as identify the type of biology as PGRMC-related for other steps. We can posit the broad hypothesis that eukaryogenesis involved a CPR bacterial cytb5MY protein somehow contributing to the establishment of communication between mitochondrial and host cell membranes. For a CPR symbiotic bacterium with a reduced genome lacking the genes to synthesize its own fatty acids [104], and which thus must acquire them from a host cell, intermembrane communication is very probably a CPR function that could have been coopted by the burgeoning eukaryote.
Thereby, in this hypothesis we expect PGRMC1 to participate in broader regulation of ER-mitochondrial communication, and thereby to be a focal point to channel cell metabolism along alternative routes that originally reflected a response to the presence of oxygen. Oxygen levels are directly related to ER stress. The formation of disulfide bonds in the ER lumen during correct protein folding is an oxidative process. Under hypoxic conditions misfolded proteins accumulate, which induces the UPR and the above-described ER stress response [377]. In eumetazoans the primary response relies on Hif-1 [381], however Hif-1 first appeared in the LEUMCA [53], and therefore superimposed its LEUMCA functions onto a preexisting system of much more ancient microbial processes.
As we have seen above, as well as having the first Hif-1 transcription factor, the LEUMCA was also the first organism to possess both the PGRMC Y139 and Y180 residues, as well as the extended C-terminus that houses the Y180 element (Fig. 8) [7]. Chen et al. [371] found that PGRMC1 directly binds to low molecular complexes targeted for ER-phagy via its C-terminal region (residues 43-195), and that a deletion of residues 171-195 (precisely the C-terminus of PGRMC1 that is novel in eumetazoans: Fig. 8) abrogated that cargo binding.
Notably for this discussion, these low molecular weight PGRMC1 cargo complexes that require the C-terminus feature small proteins such as pro-insulin, and other peptide hormones: a new class of protein for a new animal function. We saw above how the gastrulation organizer of the LEUMCA probably gave rise to pharyngeal ectoderm, which possessed insulogenic secretory cells that can be found in extant cnidarian pharyngeal ectoderm, and the mammalian pancreas [227]. Because of its different variety of cell types, the LEUMCA was the first organism to develop peptide hormone inter-cellular signaling, and also the first to develop the PGRMC1 protein C-terminal region now known to be responsible for the new unfolded protein response associated with them. PGRMC1 is also associated with the subcellular translocation to the plasma membrane of a variety of hormone receptors (for review: [47]). A new type of high abundance protein being translated in the ER and an associated new ER-phagy cargo-recruitment region of PGRMC arose in evolution simultaneously.
The fact that our hypothesis associates PGRMC/MAPR proteins with an ancestral eukaryotic oxygen response and biochemical switch function, and hence PGRMC was already sensitive and to responding to oxygen levels, sits well with the likelihood that evolution of the LEUMCA would use the PGRMC protein to deal with these evolutionarily novel hormones if they are misfolded due to hypoxia. Although the relationship between Hif-1 and PGRMC does not present itself immediately from the hypothesis, it is a strong prediction that a relationship exists and awaits discovery.
Finally, to emphasize the obvious, the insulin response that seems to have appeared newly in the LEUMCA evokes just exactly the type of overall metabolic switch which we propose the ancestral MAPR protein was associated with during eukaryogenesis. It regulates the switch between mitochondrial oxidative phosphorylation and anaerobic glycolysis, or between the catabolism of lipid alkyl carbon skeletons and the production of acetyl-CoA for their synthesis.
While ctenophores, poriferans and placozoans have paraHox genes, cnidarians and bilaterians possess Hox genes which specify anterior-posterior identity during embryogenesis. There is no evidence for a Hox cluster in cnidaria. Hox clusters evolved in bilaterian animals, and exhibit the principle of spatial collinearity, where their expression along the anterior-posterior axis of the embryo corresponds to the order of genes within the cluster [232]. There is no apparent link between PGRMC and this LEUMCA innovation, but the hox cluster was instrumental in the subsequent evolution of bilaterians.
The next major evolutionary development on the path to humanity after the LEUMCA was the evolution of bilaterian body grade by the common ancestor of bilaterian animals (the urbilaterian), with its third embryological mesoderm germ layer (Fig. 11E–G). There is obviously much more involved in bilaterian evolution than just putative effects of PGRMC (Once more analogously to most of English being of Germanic origin, but with additional influence from other languages as iterated in the abstract [119]).
For instance, unlike the LEUMCA the urbilaterian possessed all 11 classes of animal homeobox domain transcription factors [382], including a hox-cluster of at least seven genes whose expression during embryology follows their order on the chromosome [232]. Indeed, Heger et al. [54] identified 157 genes that are unique to bilaterians relative to cnidarians, including factors involved in the nodal pathways of mesoderm development, more sophisticated capacity for cell communication via an expanded G protein-coupled receptor repertoire, and components required for nervous system development and neural guidance. Doubtless, each of those 157 new bilaterian genes probably has its own compelling story. I am proposing here that PGRMC phosphorylation status (which was not among the reported 157) established an important part of the platform upon which the subsequent evolution of eumetazoan morphological diversity was built. Many of those 157 bilaterian stories may have begun with the evolutionary potential created by PGRMC Y139 and Y180 and the extended C-terminus in the LEUMCA.
The urbilaterian modified the gastrulation process to induce epithelial cells at the lip of the gastrula to undergo EMT, adopting amoeboid migratory ability which the cells use to migrate into the cavity between endoderm and ectoderm (Fig. 11E). These animals are called triploblast because of their three germ layers, including ectoderm, endoderm, and mesoderm. Whatever advantage was initially provided by formerly endodermal cells migrating to the interior following EMT, the subsequent evolution of mesoderm provided the evolutionary raw material for the later-evolving embryological process of tissue differentiation that gave rise to bilaterian body plans, with their diverse cell and tissue types. The two main triploblastic groups are protostomes (including insects and nematodes) where the gastrulation blastopore forms the mouth, and deuterostomes (which includes our branch of chordates) where the blastopore forms the anus. A third triploblastic group, the Xenacoelomorpha, are marine worms that form an early-branching sister group to the other two bilaterian groups [338, 383], however that very interesting detail is beyond the scope of this current work.
In bilaterians, the proto-mesodermal cells undergo EMT under the influence of
the organizer, changing from epithelial cells to motile ones associated with the
upregulation of N-cadherin (neural-cadherin, cadherin-2) followed by the
downregulation of E-cadherin (epithelial cadherin, cadherin-1): i.e., the
modulation of epithelial adherens junction functions. Vinculin is a marker for
EMT, which helps link adherens junctions to the actomyosin network [384]. As
noted above, PGRMC1 modulates vinculin abundance [50] and the actin cytoskeleton
[50, 91, 172], as well as
PGRMC Y139 and Y180 arose in the LEUMCA, whereas protostome and deuterostome
organisms sampled (with few exceptions) additionally possessed equivalents to
presumed regulatory phosphorylation sites T178 and S181 [7]. An NCBI BLASTp
search of 775 available Xenacoelomorpha (taxid:1312402) sequences using human
PGRMC1 (sp
The above has already discussed much about bilaterian animals. What is clear is that the animals of the Ediacaran fauna (Fig. 8A,C), the earliest fossilized animals from about 600 mya, already included triploblastic bilaterian animals, and therefore both the LEUMCA and its descendant the urbilaterian lived long before this time. Although the 95% confidence interval bars on individual estimates for phylogenetic divergences made by different studies cited by Sperling and Stockey spanned many millions of years [174], the consensus from those studies places the LEUMCA during the Sturtian glaciation, and the urbilaterian near its end (Fig. 8A).
As discussed above, the Sturtian was a time of low oxygen availability, and therefore the early evolution of animals quite possibly involved a response to low oxygen [174]. That this was an issue subject to selective advantage is attested by the acquisition of the oxygen-responsive transcription factor Hif-1 by the last common ancestor of placozoans and the LEUMCA [53, 212]. That same organism also acquired a brand-new C-terminal extension of the PGRMC1 gene [7], suggestive of related functions between PGRMC1 and Hif-1. By the time of the LEUMCA (if indeed placozoans are not descended from the LEUMCA), that C-terminal extension had been garlanded by Y180, and the MIHIR had acquired Y139.
This proposed biological relationship may be retained in the biology of Hif-1 and PGRMC1 in humans. In the hypoxic zone of human ductal carcinoma in situ breast tumors, next to the necrotic zone, PGRMC1 was induced in some cells before the Hif-1-induced GLUT1 glucose transporter that is associated with cancer-typical oxygen-independent glycolytic metabolism, causing us back then to predict involvement of PGRMC1 in the Warburg effect [46]. Recent results show that PGRMC1 does indeed change the degree of Warburg/glycolytic metabolism [49, 50, 306]. The associated regulation of mitochondrial oxygen consumption may reflect the exploitation on the path to multicellularity of the ancient eukaryotic mechanism of mitochondrial regulation that is here proposed to be involved in eukaryogenic manipulation of host and endosymbiont by the ancestral CPR cytb5MY protein, which had become the first MIHIR-containing MAPR protein in early eukaryotes.
In this context it is useful to keep in focus the relatively primitive state of body plan of the urbilaterian organism, which presumably resembled an elongated worm- or slug-like organism (Fig. 11F,G). Importantly, the urbilaterian had developed a Hox cluster, a group of at least seven tandem Hox genes which arose by gene duplications at a single chromosome region, and where the order of genes on the chromosome dictates the order of their expression during embryogenesis. Hox transcription factors can be viewed as keys which unlock access to specific regions of the genome required for tissue-specific expression. In all bilaterians, but not cnidarians, Hox1 typically determines anterior embryonic identity, while Hox9-13 specify posterior identity. It is thought that Hox proteins of the urbilaterian originally may have specified the polarity of neurectoderm (reviewed by [232]). Epigenetic CpG DNA methylation forms a part of this system in some but not all bilaterians (e.g., nematodes and dipteran insects do not exhibit CpG methylation). During this embryological process, the most ancient genes are the ones expressed first, with endodermal genes being expressed earliest. The endodermal transcriptome most resembles that of our choanoflagellate single celled sister group [233, 234].
The urbilaterian would almost certainly be totally unviable today, but had a somewhat centralized sensory nerve ganglia, perhaps near the mouth end of its body, a digestive cavity (probably already colonized by symbiotic bacteria), contractile cells, gonads, and was maybe starting to develop specialized internal mesodermal cells, to perhaps assist with digestion, locomotion, or even oxygen transport [195, 246, 247]. However, the colonization of the embryonic blastocoel by mesoderm cells to enable the formation of connective tissues, or mesenchyme, with specialized extracellular matrices, opened up wholly new engineering possibilities for evolution to form novel body structures [195].
The resultant more complex bilaterian body plans surpassed the ability of oxygen
to diffuse
Consistent with the hypothesis proposed here, that PGRMC1 tyrosine phosphorylation affects migratory and metabolic properties during EMT, while this manuscript was under review Schwager et al. [388] reported that the glycolytic metabolism of breast cancer cells switches between migratory mesenchymal and sessile epithelial phenotypes. They identified that the increased glycolysis of migratory mesenchymal-like cells can be plastically manipulated to direct cells into dependence on mitochondrial respiration and adoption of epithelial-like phenotype, and vice versa [388]. They cited our study showing that PGRMC1 Y180 directed precisely this biology [50], but did not assay Y180 phosphorylation status. A testable prediction is that Y180 phosphorylation status will vary between migratory mesenchymal (phosphorylated Y180) and sessile epithelial (unphosphorylated Y180) cell morphotypes. That observation would greatly strengthen the hypothesis that the evolutionary acquisition of PGRMC tyrosine phosphorylation enabled the novel evolution of the gastrulation organizer in the LEUMCA. We could posit that under low oxygen tension cells may have been ancestrally predisposed to migrate to seek more oxygenated environments, and that organizer activity manipulated this predisposition in the evolutionary appearance of PSCs and the appearance of a mechanism to dynamically alter genomic plasticity.
PGRMC effects on heme and hemoglobin biology could be involved in the origins of the vasculature, considering PGRMC’s heme trafficking role [82]. Globin genes are present in organisms from bacteria to animals, including diverse holozoan protists related to animals, and so were naturally present in the LEUMCA, but underwent dramatic expansion during evolution from the urbilaterian to humans [389, 390, 391]. Although the evolution of animal oxygen transport systems is relevant to our topic, it exceeds the bounds of this work.
The suite of functions that apparently first evolved in axonal guidance in the LEUMCA, taking the Netrin/DCC ligand receptor pair as an example, has been rehashed in multiple bilaterian guidance systems. Knowing that PGRMC is involved in axon migration via an interaction with members of the DCC family from nematodes to mammals [220, 314, 315], we could hypothesize that PGRMC may be involved in the initial migration of individual epithelial cell mesodermal precursors into the embryo, or in the centralization of the nervous system.
In lactating mammals, P4 induces development of the mammary duct system whereby terminal end buds extend mammary ductules into surrounding mesenchymal tissue. This process requires PGRMC1 [359], as well as neogenin (the second human DCC family member, also involved in netrin-mediated axon guidance) in the basal epithelial interacting with netrin [392]. Since the growth cones of vascular vessels are also directed by members of the DCC family [393, 394] we could additionally hypothesize that PGRMC is directly involved in angiogenesis (for a recent review on the role of progestogens in neovascularization, see [395]), or perhaps with embryological formation of the anus. However, this is speculative and further experimental work is clearly required to establish the conserved roles of PGRMC1 tyrosines in this process, if any.
The underlying point to be noted here is that the machinery originally required for axon migration and synapse formation has perhaps been co-opted during evolution to facilitate various types of guided cell migration. It is highly probable that PGRMC tyrosine phosphorylation is involved in many such early embryological process. Furthermore, if we can imagine that actomyosin-mediated migration of an axon mechanistically resembles the leading-edge migration of a proto-mesodermal cell following EMT, then PGRMC’s role could be to reorganize the actin cytoskeleton (in a steroid-dependent manner?), including motility and cell surface localization of guidance receptors from EMT, axon guidance, vascular migration, mammary ductule formation, and more. This, once more, is hypothesis.
Finally, it has been suggested that the Cambrian explosion was facilitated by a new ability of animals to experience associative learning [396, 397], and that this may be associated with epigenetic mechanisms of cell memory [398]. In animals with complex anatomical body plans, such mechanisms extend beyond the regulation of gene expression, also including heritable influences of regulated membrane ion channel activity and cytoskeletal organization, which together are capable of considerable developmental plasticity via a kind of higher-level morphogenetic master regulator. I.e., the response to a given pattern of gene expression depends upon the pre-existing state of the cell. It remains unclear how the extra-genomic information at this level is encoded [399, 400, 401].
The acquisition of associative learning was dependent upon the evolution of the nervous systems being considered here in bilaterian organisms as diverse as mollusks, arthropods and vertebrates [402]. We will see in the accompanying paper [2] that PGRMC is implicated in the biology of both learning and epigenetics. With its ability to influence cytoskeleton, gene expression, and membrane properties, its proposed gastrulation organizer role may be related to the mechanism of extra-genomic reprogrammable higher level master regulator circuits. These are reported to influence embryogenesis, regeneration, and cancer [401].
At the outset of this section, it must be strongly stated that the author is unaware of any evidence that associates PGRMC1 with germline segregation. This section is being included because PGRMC1 is related to steroid responses of the mammalian male and female reproductive systems (i.e., germline cells), and because germline segregation appeared around the time that PGRMC1 acquired its phosphorylated tyrosines Y139 and Y180 in the LEUMCA, with especially the adjacent T178 and S181 regulatory residues in bilaterians that undergo germline segregation.
Very briefly, impregnation of the oocyte by the sperm forms the zygote. As soon as maternal genes are switched off and embryonic transcription commences, epigenomic reprogramming occurs, removing methyl groups from histones and DNA, to generate a hypomethylated ‘clean slate’ epigenetic state. The resulting cells are said to be in a “naïve” state of pluripotency and are called naïve PSCs. These can transition through a variety of epigenetic and metabolic intermediate pluripotential states, until they undergo genomic hypermethylation to form “primed” PSCs, which can then differentiate into multiple lineages. Primordial germ cells differentiate from one of the intermediate pluripotency states. Great advances in our understanding of this process at a molecular level have been made over recent years, to the extent that the process can be directionally steered in vitro to generate fully functional oocytes or spermatogonia [403, 404, 405].
This paper hypothesizes that altered actin cytoskeletal activity associated with the gastrulation organizer was enabled by the acquisitions of PGRMC1 Y139 and the C-terminal extension containing Y180 by the LEUMCA. Based upon the situation in cnidarians, the LEUMCA maintained a population of stem cells into adult life which could differentiate into either soma or germline cells, as is also the case in sponges (poriferans), and which therefore probably represented a primitive state.
In contrast to cnidarians, bilaterians develop a segregated germline early in embryology. The urbilaterian apparently developed segregation of germline cells [406, 407, 408]. Bilaterians exhibit a variety in the specification of committed germline cells during embryology, however the process involves profound epigenetic changes associated with altered genomic three-dimensional organization [177, 409, 410, 411]. The demonstration that transcription factor Activating Enhancer-Binding Protein 2 (AP2) drives this process in cnidarians and mammals [412] is compatible with bilaterians already having inherited a common eumetazoan genetic toolset for germline specification, which was used to solve the problem differently in various bilaterian lineages. We have seen a similar concept for the independent evolution of cnidarian and bilaterian striated muscle from common inherited apparatus of the LEUMCA [298], and a proposed similar independent evolution of central nerve cords from an urbilaterian nerve net [242].
Model organisms can help us to understand the basic processes involved, noting that not all processes can be directly extrapolated to mammals. The process of germline segregation in bilaterian Drosophila requires actin-dependent processes not only by future somatic feeder cells but is dependent upon actomyosin forces in the cytoplasm germline cells [413, 414] (to use the simplified terminology of this present work: see the original works for correct technical terminology). The hypothesis being forwarded here predicts that transient alterations in Y139/Y180 phosphorylation will be involved in germline segregation, as well as probably regulatory contributions by T178 and S181 phosphorylation in bilaterians, and that these very early embryological features will be largely conserved between bilaterians. They may involve modulation of the actomyosin system.
Meiosis and sex are ancient in eukaryotes; however, the production of gametes (ova in females and spermatozoa in males) is an animal innovation. Poriferans possess a suite of Germline Multipotency Program (GMP) genes that is shared with eumetazoans [210, 415]. GMP genes as currently defined do not include PGRMC, but since one of the functions activated by oogenesis is fatty acid synthesis then PGRMC could conceivably be involved.
Poriferans contain three main cell types: Choanocytes are flagellated feeding cells found in the lining of inner channels. Pinacocytes are epithelial cells lining the exterior and the inner channels. Both are derived from archeocytes, the third main cell type, and can interconvert back to archeocytes. These exhibit similarities to pluripotent stem cells of higher animals in both transcriptional profile and cell reversible plasticity [197]. Archeocytes can also produce gametes [415].
Poriferans and ctenophores do not contain a reduced set of GMP compared with the LEUMCA clade. Multiple cell types in poriferans express GMP genes and may exhibit persistent multi/pluripotency [210]. Therefore, animal germline specification was constructed using a toolkit inherited from our first animal ancestors. However, we are interested here in germline segregation.
Sponges and corals possess no differentially segregated germline cells [408], whereas other cnidarians do [410]. Interestingly, placozoans have genes for meiosis, but none has ever been observed [416]. The LEUMCA may have had germ stem cells, but the possibility remains that some cnidarians may have acquired dedicated germline stem cells independently [410]. The observation that germ cell induction is mediated by transcription factor AP2 from cnidarian Hydra to mammals [412] is supportive that this function was present in the LEUMCA and was lost in corals.
The reason for the evolutionary appearance of germline segregation has been the subject of much speculation (for discussions, see: [417, 418]). One theory proposes that metabolically inactive cells form germline to protect the genome, which is related to mitochondrial activity. In its simplest form, somatic cells are metabolically active and acquire mutations, and therefore are unsuited for germline (see [408] for arguments). Production of sperm throughout life leads to very high numbers of cell divisions. Testes have a high metabolic rate, and most human genome mutation arises from the male germline [419].
The development of dedicated germline within a body of somatic cells has been modelled as being a means to protect germ cells, even to the extent that advantages conferred by the input of information from multiple cell types with specialized functions extending towards the cognitive or conceptual level (independently of neurons). Here, stem or germ cells act as both regulatory controllers of somatic cells (“imperial masters”), and receivers of information from them. In this model the evolution of the nervous system becomes an extension of this cognitive function, with the prediction that the nervous system should be involved in the morphogenesis of the somatic body, as an extension of germ and stem cell hierarchy over somatic cells [417].
Tverskoi et al. [418] extended previous theoretical models of germ-soma specialization to include a response to environmental quality. They argued that resource restrictions could strongly influence the emergence of specialized soma and germline cell populations. That concept could be highly relevant to the origins of bilaterians around the end of the Sturtian glaciation (Fig. 8).
Sperm are motile, with multiple active mitochondria and a high metabolism-induced mutation rate in mitochondrial DNA. It has been proposed that this led to the origin of an oocyte with many metabolically quiescent mitochondria, and inheritance of mitochondrial DNA from the female lineage [408]. The argument is further developed that selection for mitochondrial quality may have underpinned the sequestration of separate germline cells in bilaterians. The concept is that there is a kind of Goldilocks zone between the ideal low number of embryogenic cell divisions that can generate advantageous mutations as a source of genetic variance, and too many cell divisions that burden mitochondria with excessive mutational loads. The hypothetical solution was to partition the germline from somatic cells early in embryogenesis in organisms with more complex body plans. However, the argument is more detailed, and the reader is referred to the original source for particulars [408].
An alternative (but not exclusively so) proposal is the “dirty work hypothesis”. Here, somatic cells evolved to permit cells to perform functions which would be too detrimental to the genomic integrity of a germline cell, and which may be related to the evolution of aging in animals [420].
Germline cells are typically maintained by specialized somatic feeder cells. PGRMC1 is expressed in the granulosa [22] and Sertoli [355, 356] nurse/feeder cells of female and male germline cells respectively in mammals.
BMPs direct germ cell lineage in chordates but not cnidarians, and so are unlikely to involve an urbilaterian organizer-related PGRMC function (unless BMP-signaling is subsequently discovered to drive or previously have driven cnidarian germ specification). P4 secretion by granulosa cells is suppressed by BMP ligands, whereas BMP15 exerts no response [421].
BMP15 and growth/differentiation factor 9 (GDF9, another BMP member) are secreted by ovaries to support follicular growth [422]. PGRMC1 is one of several proteins known genetically to be expressed in granulosa cells and involved in failure of ovarian development (primary ovarian insufficiency: POI), and therefore to be crucial in follicular maturation [22, 144, 358, 423, 424, 425, 426, 427, 428]. PGRMC2, which diverged from PGRMC1 during early chordate evolution [6, 12] serves a related function [429]. Therefore, PGRMC biology seems intimately associated with the female germline.
As an aside, PGRMC1 is implicated in the P4-dependent response of the sperm acrosome reaction [275, 430]. For a review, see Thomas [267]. The acrosome reaction is initiated when the sperm encounters P4 in the jelly surrounding the oocyte and is the process whereby sperm is endocytosed by the oocyte, releasing the haploid genome into the oocyte cytoplasm. However, the acrosome reaction developed in eutherian mammals as a specialization related to internal fertilization, and so is not immediately relevant to bilaterian origins [431, 432].
Otherwise, there is no direct evidence that PGRMC is involved in germline establishment. Germline defects were not reported in the knockout of the nematode C. elegans PGRMC1 homolog (vem1) [315], and germline developed in mouse [163], and zebrafish [26, 273] knockouts (See Caveat Emptor Section 10, below). PGRMC1-regulated steroidogenesis is required for normal fertility in the latter. However, if we associated PGRMC1 with Warburg glycolytic metabolism then a new set of circumstantial evidence presents itself.
Sertoli cells, the male gamete feeder cells of mammals, exhibit Warburg aerobic glycolysis which produces lactate that is required for spermatogenesis. Sertoli cells perform a host of essential functions, forming a blood-testes barrier which immunologically shields germ cells from the rest of the body. Lactate, the product of anaerobic glycolysis, performs an essential role in spermatogenesis [433]. Germ cells take up lactate from Sertoli cells, reduce it to pyruvate, and this was thought to be utilized for ATP production via the TCA cycle [434]. However, the reduction of lactate to pyruvate produces cytoplasmic NADH which is used to produce reactive oxygen species by NAD(P)H oxidase 4, and the ROS are thought to act as second messengers that regulate spermatogenic pathways in germ cells [435]. It will be interesting to see whether this oxidative induction of meiosis is in any way related to a eukaryogenic origin of meiosis.
The oocyte female germ cells, which require active pentose phosphate pathway for meiosis, rely on pyruvate and not glucose to generate TCA substrates, which they are fed by follicle cells (mainly granulosa nurse cells). These actively metabolize glucose to pyruvate via glycolysis between mid-preantral and preovulatory stages [436]. Not expressing the classical PGR, granulosa cells are the classic example of a cell type where P4 response is mediated by PGRMC1 [13, 14, 15, 22, 437]. PGRMC1 mediates a P4-dependent induction of Warburg metabolism in HEK293 cells [49] and neurons [306]. The work of Peluso, Pru, and colleagues in characterizing PGRMC1 in the granulosa cell system deserves special recognition in the field of PGRMC1/2 biology and reproductive tissues [22, 57, 182, 358]. Recently, PGRMC1 was one of a number of genes downregulated in association with placentitis, an inflammation of the placenta that causes impaired fetal development, premature birth, or abortion [438], consistent with PGRMC1’s important role in the female reproductive tract. For a recent review on P4 receptors in the female reproductive tract, see Medina-Laver et al. [439].
Newman [195] reviews how differentiated animal cell specializations probably co-opted suites of genes that had been co-regulated in their unicellular ancestors. In single celled organisms, histone modifications had been utilized to elevate or suppress gene expression since the first eukaryotes. Metazoans including poriferans invented upstream or downstream enhancer regions whose transcription factor binding sites resembled those of gene promoters proximal to transcription start sites, but which could regulate the expression levels of large regions of chromosome [195].
Enhancers permitted cells to switch on or off parts of the chromosome so that cells with a single genome could exhibit totally different phenotypes [195]. Bilaterian embryology is associated with substantial changes in the three-dimensional organization of the genome because of epigenetic changes [411, 440].
Transcriptionally active gene activity can be up- or down-regulated by modulated protein binding to associated transcriptional enhancers, which are cis-acting genomic sequences with specific transcription factor binding sites [441]. Enhancer activity, especially in vertebrates, is regulated by methylation of cytosine residues adjacent to guanosine in the DNA sequence at so-called CpG (cytosine-phosphodiester-guanosine) motifs. In vertebrates CpG motifs have been negatively selected and are largely located in “islands” found in promoters and to a lesser extent enhancers [442]. DNA methylation at CpG motifs is ancient in eukaryotes, although it can be absent from specific species, such as the yeast S. cerevisiae, and the nematode C. elegans [443].
While CpG methylation of promoter and enhancer elements is most often associated with transcriptional silencing, demethylation itself is not sufficient for transcriptional activity, and the rules related to individual enhancer activity remain poorly understood [409]. Once this mechanism existed in the ancestor of the LEUMCA, the fuel for eumetazoan evolutionary diversity was available, apparently awaiting only the acquisition of a gastrulation organizer, lineage determining transcription factors, novel tissue-specific proteins, and sufficient time for natural selection to act to produce the hierarchical cascade such as vertebrate embryogenesis (or perhaps more incredibly, the larval, pupal metamorphosis, and adult life cycle stages of insects).
The rest, as they say, is evolutionary history. However, this is relevant to PGRMC phosphorylation because this process is/was initiated by the gastrulation organizer (which coincided evolutionarily with the acquisition of PGRMC1 Y139 and Y181), and mutation of PGRMC1 phosphorylation sites causes dramatic changes in genomic CpG methylation state of cancer cells [19]. Gastrulation itself requires enhancer demethylation in mice [444], and thousands of enhancers are coordinately demethylated during early vertebrate embryogenesis in the phylotypic stage, the early embryogenic stage when different taxa are morphologically similar [445].
Primed induced PSC (iPSC) formation from somatic cells involves a net increase
in the level of genomic CpG, accompanied by the active demethylation of specific
pluripotency-associated enhancers [409]. Mouse embryonic stem cells cultured in
media along with feeder cells, or supplemented with leukemia inhibitory factor
(LIF), exhibit similar hypermethylation of the genome. However, culturing
embryonic stem cells in serum free media that contains two kinase
inhibitors—targeting mitogen-activated protein kinase kinase (MEK) and
GSK-3
Note here that pluripotent embryonic stem cells represent the pre-gastrulation stage of development, since they can be induced to form gastrulating embryoid bodies [449, 450]. When we mutated PGRMC1 Y180 [19], whose evolutionary appearance coincided with the appearance of gastrulation [7], the cells superficially converted to a hypermethylated state resembling the genomic hypermethylation level of primed embryonic stem cells, representing the pre-gastrulation blastula. This is reminiscent of the report from David Sinclair and colleagues that ectopic expression of transcription factors Oct4, Sox2 and Klf4 in retinal ganglion cells led to a restoration of youthful DNA methylation patterns, which was accompanied by a revitalization of neural functions [451]. These are the three of the same transcription factors previously shown by Takahashi and Yamanaka to revert somatic fibroblasts to iPSCs [452]. The novelty of the Sinclair study’s findings was that gene dosage can induce partial reversion from sub-optimal/diseased state back to correctly differentiated state, without promoting total dedifferentiation and stem cell characteristics with associated risks such as increased cancer propensity [451]. The author predicts that Oct4, Sox2 and Klf4 will cause PGRMC1 post-translational status as part of their reprogramming mechanism of action.
In line with the hypothesis that the pleiotropic effects of PGRMC1 on cell differentiation state are related to epigenetics and genomic status, Pedroza et al. [453] profiled the miRNome of MDA-MB-468 triple negative breast cancer cells after AG-205 treatment to inhibit PGRMC1 function (although they do not acknowledge that AG-205 has off-target effects [90], see Section 2.1.1 above) or anti-PGRMC1 siRNA transfection to reduce PGRMC1 levels.
This was the first study to compare attenuation of PGRMC1 with the effects of AG-205 treatment. Although the miRNAs most strongly altered by AG-205 and PGRMC1 siRNA were non-overlapping (1008 miRNAs following AG-205 treatment and 776 miRNAs after PGRMC1 siRNA), Pedroza et al. [453] discussed the results broadly under PGRMC1 effects, with the clear assumption that all AG-205 effects are mediated through PGRMC1. For instance, in the results description of AG-205 treatment, the section heading begins with “PGRMC1 Signal Disruption Alters miRNAs…”. The discussion section makes no discrimination between AG-205 and siRNA PGRMC1, conflating both sets of results into one dialogue. From the results descriptions, Kyoto Encyclopedia of Genes and Genomes (KEGG) terms of most affected miRNA-affected pathways for AG-205 and PGRMC1 siRNA treatment involved p53 signaling pathway, cell cycle and pathways in cancers, albeit via different miRNAs and affected genes [453]. There is no data presented which shows that AG-205 exerts only PGRMC1-specific effects, and non-specific effects were not considered by the authors. The degree to which AG-205 effects are due to PGRMC1 requires urgent evaluation.
More interestingly in the context of the current discussion, by considering only the PGRMC1 siRNA results, we can conclude that the presence of PGRMC1 is required for the persistent expression status of hundreds of regulatory miRNAs (to either maintain or repress transcription, respectively). One way in which that could be accomplished would be by PGRMC1 participation in a mechanism responsible for epigenetic genomic maintenance, as we observed in MIA PaCa-2 pancreatic cancer cells expressing PGRMC1 phosphorylation site mutants [19]. Pathways associated with cell cycle and cancers can be considered as surrogates for dedifferentiation. This is a testable hypothesis, which predicts that the AG-205 and PGRMC1 siRNA treatments will exhibit induced epigenetic changes in the MDA-MB-468 cell system of Pedroza et al. [454].
In this respect, the report of Kim et al. [140] that PGRMC1 maintains
primed PSC pluripotency seems extremely relevant. They reported that PGRMC1
knockdown inhibited GSK-3
A previous study by Sperber et al. [455] had reported that stem cell pluripotency was maintained by the production of the metabolite 1-methylnicotinamide (1-MNA) by the enzyme nicotinamide-N-methyltransferase transferase (NMNT). They proposed that consumption of S-adenosyl methionine (SAM) in this reaction pleiotropically reduced the levels of available SAM methyl donor, and so pleiotropically the level of histone methylation, leading to the induction of the Wnt pathway [455]. Note that Kim et al. [140] concluded that stem cell pluripotency of primed PSCs was affected by PGRMC1, while Sperber et al. [455] concluded that NNMT activity maintained the naïve state. We found that PGRMC1 Y180 phosphorylation status modulated NNMT levels, but we observed no effects on SAM levels while we observed altered genomic CpG methylation [19]. While our study was not performed in stem cells, the pathways involved are consistent with PGRMC1 potentially being responsible for modulating the genomic epigenetic landscape of pre- and post-gastrulation cells. This theme is discussed in terms of aging biology in the accompanying paper [2].
Finally, the seemingly unrelated biology of sumoylated PGRMC1 binding the
TCF/Lef sites [142, 143, 144], GSK-3
As the mammalian embryo progresses from zygote to blastula, and begins to prepare for gastrulation, the parental CpG methylation pattern is reset to generate a hypomethylated state that corresponds to naïve PSCs. This progresses to the hypermethylated state of primed PSCs around the time of gastrulation (E6.5/E7.5 in the mouse). While most CpG sites in the genome are methylated, a relatively smaller number of sites are hypomethylated. A number of these hypomethylated sites become progressively methylated during the lifespan, which can be used to measure biological age [456]. This mechanism, which has become known as the Horvath clock, is conserved across mammalian species [457, 458, 459].
Of extreme relevance to this section, the aging clock is reset to zero around the time of gastrulation. Cultured PSCs do not age, and the methylation clock begins once primed PSCs begin to differentiate, or when differentiation processes are induced by gastrulation [460]. PGRMC1 obtained its tyrosines in evolution as the same time as the gastrulation organizer [7], and mutation of Y180 in a cancer cell line induced a hypermethylated state reminiscent of primed PSCs [19]. The primed type of PSCs most probably evolved with the LEUMCA (PGRMC1 Y139 and Y180), or the urbilaterian (PGRMC1 T178 and S181), at a time when more highly differentiated cell types did not yet exist, and the metabolic switch was related to some aspect of the survival of an animal that was relatively simple by modern standards.
The fetal membrane, or amniochorion, is a composite membrane formed by fusion of the amnion and chorion. In amniotes (land vertebrates, excluding amphibians) the fetus develops within the amniochorion. No clearly intermediate structures are known among amphibians. The fetal membrane is a classical synapomorphy for amniotes, and a major evolutionary innovation that permitted vertebrate colonization of the terrestrial surface [461].
During gestation, P4 maintains an epithelial state of amniochorion cells, which express PGRMC1 and PGRMC2, but not the classical nuclear steroid P4 receptors PGR-A (96 kDa) or PGR-B (116 kDa). Lozovyy et al. [462] showed that fetal membrane integrity during gestation is maintained via P4 acting through both PGRMC proteins.
We have previously reported that the proline-rich PGRMC1 62-66 SH3 target motif (Fig. 1B) is shared by amniotes but not amphibians and speculated that PGRMC1 may serve a role in the amniotic egg [6, 7]. We also identified a potential RGD integrin-interaction site at PGRMC1 47-49 which is present in cartilaginous and bony fish, amphibians, some reptiles, and placental mammals. It was absent in turtles, crocodilians, birds, and marsupial mammals [7]. Interestingly, these groups all exhibit major evolutionary changes in egg gestation which characterize their respective phylogenies.
The RGD motif could only be functional if PGRMC1 was present as a type 2 membrane protein (lumenal C-terminus), as opposed to a type 1 protein (cytoplasmic C-terminus). PGRMC1 has been reported to exhibit type 2 topology in neural [64, 373, 374] and cancer cell [375] settings. We speculated about the putative RGD motif that: “it is conceivable that this motif is involved in the evolution of post-fertilization vertebrate embryology, perhaps involving the relationship between amniotic sac and eggshell, or other major features related to differences in vertebrate embryology and oocyte/egg biology between these groups” [7]. The finding that PGRMC1 regulates the gestational integrity of the amniochorion [462] suggests that RGD motif and/or the SH3 target motif and its adjacent presumed S54 and S57 regulatory phosphorylation sites [5, 6] may be involved. This hypothesis requires experimental validation.
Interestingly, S54 is conserved across placental mammals (among species sampled) while S57 is restricted to primates [6, 7]. S54 can also be linked to chondroitin sulfate, a glycosaminoglycan polysaccharide of the extracellular matrix, identifying PGRMC1 as a proteoglycan core protein [463]. In the folded PGRMC1 protein, these residues are proximal to both the 47-49 RGD motif and the 62-66 SH3-target motif. Furthermore, in the folded protein structure the N-terminal SH3 target region and the C-terminal SH2 target region centered on Y180 are adjacent to each other in three-dimensional space. e.g., the peptide backbones of F73 forms H-bonds with those of K169 and L170 (Fig. 13A, Ref. [4]), and these are within Angstroms of the SH2 target motif centered at Y139 in the MIHIR (Fig. 13B). The tyrosine 139 hydroxyl H-bonds with backbone atoms of K172 and G174 (Fig. 13C) [4, 6]. Interestingly, SH3 domain proteins are usually cytosolic, but do exist luminally where they can be involved in protein/membrane trafficking [464, 465]. Therefore, protein interactions at the SH3 target motif could regulate PGRMC1 interactions with the extracellular matrix via the RGD motif and integrins, or proteoglycans at S54 for a type 2 membrane orientation, or with the tyrosine phosphorylated SH2 target motifs centered on Y139 and Y180 for a type 1 membrane protein with access of the C-terminus to cytoplasmic kinases and phosphatases.
 Fig. 13.
Fig. 13.Spatial proximity of some PGRMC1 residues. Images were generated with the “3D view” function from https://www.rcsb.org/3d-view/4X8Y/1andlabeledmanually. (A) The peptide backbone of F73 forms H-bonds with backbone atoms of K169 and L170. (B) Residues from A are within Angstroms of Y139. The position of the peptide backbone atoms of residues is shown. The image also shows the proximity of these residues to V179 (adjacent to Y180) and S68 of structure 4X8Y at the position of PGRMC1 L68, which is adjacent to the PPPLP SH3 target sequence from 62-66. S68 is one of four non-PGRMC1 residues present in the 4X8Y PGRMC1 crystal structure, some of which directly contact heme [4]. (C) The hydroxyl group of Y139 form H-bonds with the peptide backbones of K172 and G174.
The Latin caveat emptor ‘let the buyer beware’ provides a fitting cautionary reminder of the conjectural nature of some of the theses presented above, before progressing to consider quo vadis ‘where are you going’ PGRMC in the accompanying manuscript [2]. The above builds an attractive case that PGRMC tyrosine phosphorylation was acquired at the same time as the gastrulation organizer, the Y180 module is associated with actin cytoskeletal and epigenetic effects, and a case can be made that PGRMC1 redox-related biology could be involved with the glycolytic metabolism of stem cells, as well as resonating with the hypoxically stressful environmental circumstances associated with the evolution of the gastrulation organizer of the LEUMCA, and subsequently the urbilaterian.
This may lead to predictions that PGRMC1 would be required for embryology, and
genetic knockouts would be lethal. Early knockout studies would have supported
this prediction. A floxed pgrmc1
 Fig. 14.
Fig. 14.PGRMC1 mouse germline knockout models. Top: Schematic
presentation of mouse PGRMC1 (UniProt O55022). The positions of transmembrane
peptide (TM), the PPPLP SH3 target motif (SH3), and SH2 target motifs surrounding
Y139 and Y180 [5] are indicated. Middle: The Pgrmc1
Another study with the Pgrmc1-TALEN knockout showed that PGRMC1 was involved in mammary gland development [359], and another that PGRMC1 caused a P4-induced increase in hepatic glucose synthesis in the absence of insulin [51]. Effects on fertility have not been reported in this knockout model.
Deletion of zebrafish PGRMC1 [273], PGRMC2 [274], or the double knockout [26] also resulted in development to adulthood, albeit with reduced fertility (lower number of offspring) and impaired steroidogenesis. The double knockout failed to ovulate and had impaired nuclear PGR expression. It is not clear from the zebrafish work that the authors even considered the possibility that other developmental processes (e.g., hepatic lipid or glucose homeostasis, vascularization, heme biology, mitochondrial status, oxidative stress response) could be affected, because they seem only to have assayed processes associated with fertility. Knockout of the C. elegans PGRMC1 homologue vem1 produced adults that exhibited impaired motor functions due to embryological faults in axon guidance in a subset of neurons [315].
Therefore, it appears that whatever essential function is exerted by the PGRMC1
tyrosines which were acquired and strongly (but not universally) conserved since
the evolutionary stage of the LEUMCA [7], that function is not essential for the
development of the adult body if the entire protein is absent. The strong
evolutionary conservation of PGRMC1 tyrosines Y139 and Y180 among eumetazoans
argues for a critical eumetazoan function, which may be solely related to
fertility. If there is a critical role in organizer function,
Wnt/
Following from the arguments presented above in section 7.4.7 “A one brain theory of PGRMC-associated LEUMCA intelligence”, it will be interesting to test whether PGRMC knockouts exhibit learning, behavioral, or conceptual deficits relative to wild-type animals. The accompanying paper [2] will address how the biology reconstructed above may focus our understanding of some modern pathologies.
1-MNA, 1-methylnicotinamide; A
MAC conceptualized and wrote this work.
Not applicable.
I am grateful to Tim Cahill (R.I.P.) for Latin language assistance.
This research received no external funding.
Michael A. Cahill is scientific advisor to and minor shareholder (
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
