Introduction: The ambiguity of the drug target is one of the major factors restricting the development of traditional Chinese medicine (TCMs) and its bioactive constituents. The characteristics of “multiple components, multiple targets and multiple pathways” of TCMs make the research of drug targets extremely difficult. With the emergence of new theories, there are increasing technologies and strategies that can be used for the drug targets research of TCMs. In this paper, we summarize several techniques and methods applied to the study of TCM targets. Methods: Through consulting a large number of literature, research and summary, and finally summarized the application direction of the technical method, advantages and limitations. Results: The methods and techniques including computer aided drug design, network pharmacology, phage display, affinity fishing, drug affinity responsive target stability and cellular thermal shift assay were summarized, and their application directions, advantages and disadvantages were discussed. At the same time, a large number of application examples were given to provide reference for the research of TCM targets.
Traditional Chinese medicine (TCM) is one of the precious treasures of the world. For some diseases, traditional Chinese medicine (TCM) has attracted increasing attention because of its significant efficacy and few side effects. It’s known that chemical drugs are single compounds that act on specific targets. Unlike synthetic drugs, a single TCM is composited by lots of components. In clinical, the medicine is commonly used in the form of prescriptions, and each prescription contains several TCMs. There are many ingredients in traditional Chinese medicine prescriptions, and play a therapeutic effect by affecting multiple targets of the disease. Because it does not play a therapeutic role through a single target, the incidence of drug resistance to traditional Chinese medicine is low. Chu et al. [1] studied the composition comparison between single decoction and combined decoction of Ephedra and Pheretima, and the results showed that combined decoction was better than single decoction and combined use of ephedrine and pseudoephedrine as detection indicators. The chemical components in traditional Chinese medicine will undergo aggregation, precipitation, dissolution and other reactions during the combined decoction. Some chemical components have low dissolution rate or basically cannot dissolve in single decoction, but when other components are present, their dissolution rate can be increased, thereby exerting effect. Therefore, the research of traditional Chinese medicine targets is more difficult than chemical medicine, and more work needs to be done.
At present, there are many studies on the material basis and mechanism of action of the TCM. The ultimate goal of these studies is to treat diseases using these TCMs, and the drug target is the bridge between drugs and diseases, but there are few studies on the targets of traditional Chinese medicine [2]. However, the research of drug targets is especially difficult. For a few active ingredients with clear targets, the process of target discovery in vivo is somewhat “by accident”. The most representative is the discovery of cannabinoid receptors. In the late 1980s, Matsuda et al. [3] stably transfected a cDNA (SKR6) isolated from the rat cerebral cortex cDNA library into CHO cells, and found a reaction in vitro when incubated with cannabinoids, which may be the reason for the relative lack of current target research results. There are many researches on single target and drug molecules in recent decades, but drugs with single molecular targets are often difficult to achieve the desired effect or highly toxic when treating diseases. It is difficult to cure multi gene diseases such as tumors; diseases affecting multiple tissues or cells such as diabetes [4]; diseases with more damage mechanisms such as stroke [5]. Therefore, in order to provide a reference for drug targets research of TCMs, this paper aimed to briefly introduce the applications of these strategies and technologies in drug targets exploration, which would be beneficial to the further molecular mechanism investigations of TCMs and its bioactive components.
New drug research is a complex, long and risky undertaking, which will consume a lot of manpower, material resources and time. At the same time, it is possible to fail in each stage of research and development, and eventually lead to the failure of development. New drug research and development generally includes the following stages: target confirmation, new drug discovery, preclinical evaluation, clinical trial, administrative examination and approval, post marketing supervision, etc. [6]. Furthermore, one of the most important steps in new drug research is to find possible molecular targets. Only by clarifying the drug’s mechanism of action and observing whether the drug can improve or reduce or even avoid the damage caused by the disease during the occurrence and development.
Ischemic stroke has attracted worldwide attention due to its high mortality, high disability rate and high recurrence rate. As one of the new drugs independently developed in China, n-Butylphthalide (NBP) has the effects of multi-target anti cerebral ischemia, anti-thrombosis, anti-platelet aggregation, improving microcirculation, improving blood flow of cerebral ischemia area and cerebral functional metabolism, scavenging free radicals, inhibiting oxidative stress, protecting nerve cells and inhibiting apoptosis, etc. [7, 8, 9, 10, 11, 12, 13, 14, 15]. Ren et al. [16] found that NBP can promote the expression of Mipu1 gene and play a role in reducing the degree of pathological damage in brain tissue, indicating that Mipu1 may be the target of NBP. Studies have shown that: Myocardial ischemic preconditioning up-regulated protein 1 (Mipu1) may be through down-regulating the expression of the pro-apoptotic gene Bax, while up-regulating the expression of the anti-apoptotic gene Bcl-2. And change the ratio of Bcl-2/Bax to reduce hypoxic-ischemic injury, which may be related to cell protection function [17, 18]. Yang et al. [19] used rat primary brain microvascular endothelial cells (BMECs) and neurons to co-culture and then giving them oxygen glucose deprivation (OGD). The results showed that the protective effect of NBP on neurons depended on the normal function of BMECs, indicating that the direct cellular target of brain protection of NBP was BMECs. NBP is a multi-target drug, and a single target cannot fully explain the reason for its efficacy. Therefore, the research on the target of NBP is still in progress. Only when the target of NBP is completely found can the function and toxicity of NBP be fully understood.
Ligustrazine is another new drug developed based on the traditional efficacy of commonly TCM. In the early 1970s, Chinese scientists carried out research on the effective ingredients of Ligusticum chuanxiong Hort. and successfully separated out ligustrazine [20]. The main pathological features of vascular proliferative diseases such as atherosclerosis are the subintimal migration and proliferation of vascular smooth muscle cells (VSMC). Di and Zhang studied the effect of ligustrazine injection on the signal transmission of adhesion molecules during the abnormal proliferation of rat VSMC induced by platelet-derived growth factor (PDGF). The results found that ligustrazine can reverse the abnormal proliferation of VSMC caused by PDGF stimulation, block the signal transmission of adhesion molecules, and inhibit the expression of adhesion molecules. This is a molecular target of ligustrazine for promoting blood circulation and removing blood stasis [21].
The mechanism of TCM treatment of diseases is multi-target and multi-level. For the same disease, it can achieve the effect of treatment by affecting multiple targets. Different diseases may have the same or different targets, and TCM may affect these targets to achieve the purpose of treating different diseases. The basis of new drug research is to discover the molecular targets of drugs. At present, there are still some difficulties in discovering drug targets, especially the molecular targets of TCM and its active ingredients. Therefore, this study aims to review the important research progress of target research strategies and technical methods, and provide some help for finding drug targets to reduce the difficulty of new drug research.
With the rapid development of molecular biology, X-ray crystallography and computer science, the computer aided drug design (CADD) has developed from the original basic theoretical research into a practical subject [22]. The traditional way to discover new drugs is based on a large number of compounds (including natural products and synthetic chemicals derived from animals, plants and microorganisms) through extensive screening under various activity determination models. But this method requires a huge number of manpower and material resources, and the blindness is greater and the accuracy rate is low. CADD is a drug design method that rationally designs new structural lead compounds by simulating the interaction between drugs and receptor bio-macromolecules or by analyzing the internal relationship between known drug structures and activities [23]. A few decades ago, drug targets could only be discovered through general sieve, which took a lot of time and new drug research could only be carried out at a very slow rate. With the rapid development of CADD, theoretically, as long as the computer operation speed is fast enough, through computer simulation, calculation and budget of the relationship between drugs and receptor bio-macromolecules, we can find suitable drugs or receptor molecules in tens or millions of candidate compounds. And the most suitable drug target molecules can be found through experimental verification, which greatly shortens the time for new drug research.
CADD methods are roughly divided into ligand-based drug design (LBDD) and structure-based drug design (SBDD), and a recent popular new drug design method named fragment-based drug discovery (FBDD) [24].
SBDD methods include molecular docking, new drug design, etc. Molecular docking is the most suitable technique for target discovery in computer aided drug design. The central idea of molecular docking technology originated from the “lock and key model” proposed by Fisher E. 100 years ago. Of course, the lock and key must match each other to open, but molecular docking is not exactly the same as the “lock and key model”. Molecular docking is a process in which two or more molecules recognize each other through geometric matching and energy matching [23]. The characteristics of molecular docking make it have great advantages in the discovery of new targets in traditional Chinese medicine. At present, molecular docking can be divided into forward docking and reverse docking. Forward docking is the three-dimensional conformation of a known target (Fig. 1A). By screening small molecule compound libraries, drug molecules that can stably bind to the target are found. Reverse docking is to combine the same known compound with different target proteins, screen out the target protein that can bind stably, and find the potential drug target of the drug molecule (Fig. 1B) [25]. According to whether there is a conformation change of the ligand or receptor molecule during the docking process, molecular docking can be divided into rigid docking, semi-flexible docking and flexible docking. Rigid docking, that is, during the molecular docking process, the conformation of the ligand and acceptor molecules does not change. After the molecular model is established, this method is generally used for preliminary analysis to determine whether the molecules can bind. Semi-flexible docking, that is, the conformation of the receptor is fixed during the docking process, and only allowed the ligand to have a certain extent change to adapt to the protein environment of the binding site. This docking method can improve its predictive ability while taking into account computational efficiency, making it widely used in drug design. Flexible docking, that is, during the docking process, both the receptor and the ligand molecule has a certain degree of flexibility. The ligand and the receptor will undergo conformational changes due to the mutual inducing effect, and eventually the two will adapt to each other to achieve the best binding mode. Flexible docking is more in line with the real physiological environment and has the strongest predictive ability, but it also increases the amount of calculation. Generally, this docking mode is only used in the process of precise identification between molecules [26]. Therefore, it is generally necessary to select an appropriate docking method according to the characteristics of the three docking methods before performing molecular docking. Generally, rigid docking can be selected during the initial screening of drugs to increase the speed. After selecting the appropriate drug molecules, semi-flexible or flexible docking and combination of the two were conducted to improve the predictive ability (The basic steps of molecular docking are shown in Fig. 2).
 Fig. 1.
Fig. 1.Schematic diagram of forward molecular and reverse molecular docking.
 Fig. 2.
Fig. 2.The basic steps of molecular docking.
Molecular docking is usually combined with virtual screening to screen the compound library using the computational power of the computer, so as to find the appropriate targets or drug molecules. Virtual screening is to give several keywords, such as three-dimensional structure parameters, etc., search and compare in multiple known databases, and finally find the target or the required small molecules. At the same time, molecular docking has also been applied to detect lead compounds in new drug research. Pei et al. [27] used molecular docking technology to analyze the main active ingredients and corresponding key targets of Carthamus tinctorius L. The results show that quercetin, luteolin, kaempferol, etc. have good binding activity with key targets such as AKT1, IL6, MAPK1, MAPK8, VEGFA, etc. By acting on key targets such as AKT1, IL6, VEGFA, MAPK1, MAPK8, JUN, etc., it can inhibit inflammatory response, resist oxidative stress and regulate immunity. Gu et al. [28] studied the mechanism of Indigo Naturalis in the treatment of ulcerative colitis. Through the molecular docking verification of indigo, indirubin and tryptanthrin, AHR, MAPK1 and EGFR were selected from the core targets for verification. The results showed that the docking was successful. The mechanism for the treatment of ulcerative colitis is to exert curative effect through the action of the above three components and the target (The software and characteristics of molecular docking are shown in Table 1, Ref. [29, 30, 31, 32, 33, 34, 35, 36, 37, 38]).
| Software | Feature | References |
| DOCK | The structures of ligands and receptors are known in advance and are considered rigid. | [29] |
| AutoDock | Large search space and reasonable calculation cost. | [30] |
| Affinity | The approximate location of the binding site of ligand and receptor is known. | [31] |
| InvDock | Prediction of unknown target proteins and proteins related to drug side effects and toxicity. | [32] |
| TarFisDock | Used to identify new synthetic compounds or newly isolated natural products. Identify compounds with known biological activity or targets with unknown mechanism of action. | [33] |
| Idtarget | It can screen all protein structures in PDB (protein database) and replicate the non-targets of known drugs or drug-like compounds. | [34] |
| FlexX | It can automatically calculate the molecular docking of multiple molecules in the database, and then give the best results, and the graphical interface is easy to operate. | [35] |
| GOLD | The structure of the amino acid residues around the active site must be complete, and the calculation time can be saved. | [36] |
| Surflex-Doc | Eliminate false positive results and multi-target docking. | [37] |
| LigandFit | The intrinsic defects in proteins are detected as candidate active sites, and the shape complementarity between ligands and protein active sites is considered. | [38] |
Traditional Chinese medicine has complex components and many targets. The method of using general sieve is time-consuming and labor-intensive, which is not conducive to the research of traditional Chinese medicine. Molecular docking provides convenience for the research of traditional Chinese medicine targets. After pharmacological experiments or literature review and other methods, some possible targets can be selected. Then, using the theory of molecular docking, the effective components of traditional Chinese medicine can be docked with the target, the score is calculated, and the more suitable target can be quickly screened out. The molecular docking of the effective ingredients of traditional Chinese medicine is carried out in a large number of targets, and the possible biological targets are screened out. Finally, pharmacological experimental verification or other technical verification can be used to find the target of the effective ingredients of traditional Chinese medicine and provide theoretical support for the onset of the effect of traditional Chinese medicine.
The concept of “network pharmacology” was first proposed by British pharmacologist Hopkins in 2007, and defined it as a branch of pharmacology that uses network methods to analyze the relationship between drugs, diseases and targets [39]. Network pharmacology is an emerging science based on the multi-level network of disease-gene-target-drug to predict drug targets as a whole and improve the efficiency of drug discovery. The characteristics of the multi-level network of network pharmacology coincide with the characteristics of the “multi-component, multi-target, multi-path” treatment of diseases in traditional Chinese medicine, so that network pharmacology has certain advantages in the discovery of traditional Chinese medicine targets.
The general operation mode of network pharmacology is as follows: In the first step, a series of basic information such as the structure and pharmacokinetics of required compounds are searched in relevant databases, for example, commonly used databases of TCMs and natural chemical components TCMSP [40] and TCMID [41], etc. The second step is to look for genes related to diseases, through some commonly used disease gene databases, such as GeneCards database [42], Online Mendelian Inheritance in Man (OMIM) database [43], etc. The third step is to find the target gene of the compound according to the literature screening or database query. The disease target gene and the target gene of compound interaction are intersected by the software to find the disease target gene that the compound can affect, and draw a venn diagram. The fourth step is to construct the network diagram of “drug-active ingredient-disease-target”. At present, the software of Cytoscape is generally used, and then the Network Analyzer tool built in the software is used to calculate the degree of nodes. The larger the node degree value, the more important it is in the network. After preliminary screening, the compound with a larger degree is selected as the research targets. The fifth step is to construct the key target protein-protein interaction (PPI) network. The String is a database that can search for known and predicted protein interactions, in order to better understand the relationship between target gene expression proteins PPI network diagram is necessary. Generally, PPI network is composed of multiple nodes and many edges. The larger the node, the darker the color, and the denser the connection, the more important the position of the target point. Therefore, it can be preliminarily explained that the larger the node, the stronger the correlation between the target and disease. In the sixth step, most of the research will continue to perform GO and KEGG functional enrichment analysis. The purpose of this step is to reveal the effect of the compound on the disease target gene and the pathway through which the compound acts on the target gene. GO and KEGG are two different databases. The GO database divides the functions of genes into three parts: molecular function (MF), biological process (BP), and cellular component. Molecular function (MF) refers to the activity of gene products of target genes at the molecular level. For example, Cheng et al. [44] explored the mechanism of Radix clematidis-Impatientis semen on the treatment of esophageal cancer based on network pharmacology and found that the molecular functions of key targets were mainly concentrated on protein kinase activity, phosphotransferase activity, kinase activity, etc. Biological process (BP) refers to the process that is completed through a variety of molecular activities. For example, Xu et al. [45] found that in the anti-diabetic mechanism, the active components of Rhizoma Polygonati were most closely related to the positive regulation of RNA polymerase II promoter transcription and the negative regulation of RNA polymerase II promoter transcription in biological processes. Cell composition (CC) refers to the position of the cell structure where the gene product performs its function. For example, Zheng et al. [46] used GO enrichment to analyze the mechanism of action of Curcuma longa L., and found that the cell components of target genes of Curcuma longa L. include five items, including cell junction, synapse, chloride channel complex, GABA-a receptor complex, etc. KEGG is a database based on pathways. Therefore, KEGG enrichment method is to analyze the relevant pathways of genes and reveal through which pathways compounds ultimately affect the role of genes. For example, Yuan et al. [47] did a KEGG enrichment analysis of the mechanism of action of Salvia miltiorrhiza in osteoarthritis, and the results showed that the signal pathways were mainly enriched in PI3K-Akt, IL-17, JAK-STA T, MAPK and NF-kappa B, etc. In the final step, the previously obtained active components, pathways and corresponding key targets are imported into cytoscape software to draw a network diagram of “active components–targets–pathways”, which can reflect the characteristics of “multi-components, multi-targets, and multi-pathways” of diseases treated by drugs (The basic steps of network pharmacology are shown in Fig. 3).
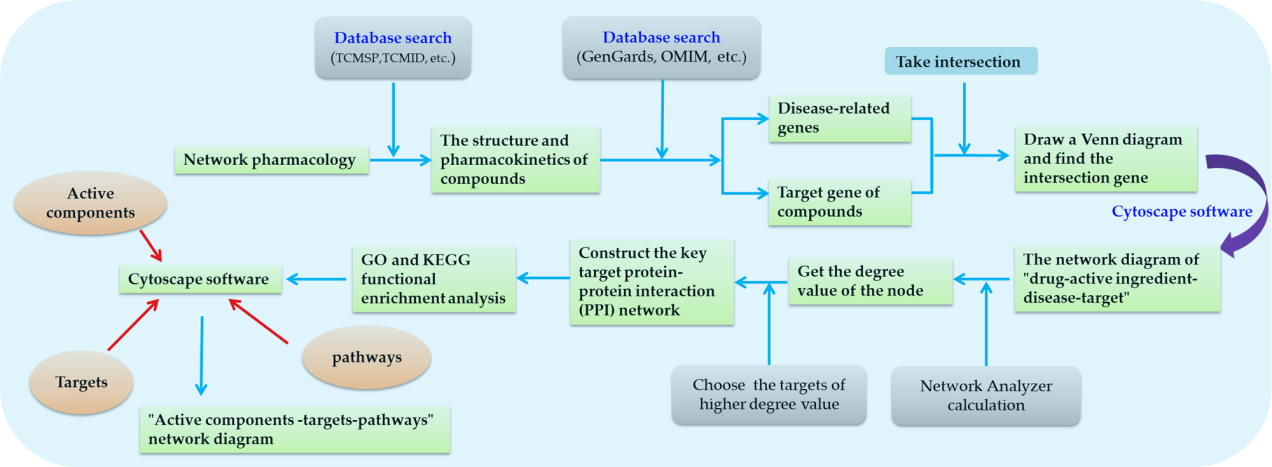 Fig. 3.
Fig. 3.The basic steps of network pharmacology.
It is appropriate to use network pharmacology to predict the target of TCM. The characteristics of “multi-component, multi-target and multi-pathway” of TCM coincide with the characteristics of network pharmacology. However, the discovery of TCM targets by network pharmacology is only a prediction without experimental verification, so it can only be used as a reference. In recent years, due to the continuous enrichment of databases, the targets discovered by network pharmacology have gradually increased, and the workload of experimental verification has also increased. Therefore, network pharmacology mostly combines with molecular docking, and further screens the binding of active ingredients to targets by using the computing power of computers. At the same time, it can preliminarily verify that molecular compounds can bind to targets, and then carry out experimental verification will greatly reduce the workload. Chen et al. [48] used network pharmacology and molecular docking to explore the mechanism of Astragalus propinquus Schischkin and Ligustri Lucidi Fructus on the treatment of Alzheimer’s disease, and screened 21 active ingredients and 41 key genes, and key targets such as AKT1, IL6, CASP3, JUN, IL1B and MAPK1, etc. The molecular docking shows that the main effective components, astragaloside IV, oleanolic acid, kaempferol, quercetin, luteolin and formononetin, could bind well to the AD (Alzheimer’s disease) marker targets AChE and NMDAR. Hu et al. [49] used molecular docking and network pharmacology method to explore the mechanism of Yupingfeng Powder in the treatment of non-small cell lung cancer, and screened out 37 active components of Yupingfeng Powder, including 146 common targets with diseases and 44 core targets. Molecular docking showed that quercetin and MAPK1, kaempferol and AKT1 had better docking activity. Both network pharmacology and molecular docking can only be used as the initial target prediction tools, but to show that the predicted target is true and effective, experimental verification is still needed.
Network pharmacology is more suitable for the initial target prediction. In the literature research, the method of network pharmacology can be used to predict the target of the Chinese medicine or the effective ingredients of the Chinese medicine to be studied. According to the predicted results, it can be combined with molecular docking or other methods for preliminary verification, and the final target can point out the direction for subsequent experiments. Therefore, in the application of network pharmacology in the research of TCM targets, although there is screening and matching of big data, the predicted targets have not been experimentally verified. To prove that the targets are true and effective, experimental verification is still needed.
Phage display technology was originally established by Smith of the University of Missouri in 1985. At that time, they linked the foreign gene fragment with gene Ⅲ(G3) of phage fd-tet, and found that the polypeptide encoded by the foreign gene fragment could be fused with the coding protein of phage gene and displayed on the surface of phage [50]. At the same time, the proteins encoded by the exogenous gene fragments displayed on the surface of phage also maintain the original three-dimensional structure and biological activity [51, 52]. The basic principle of phage display technology is: after fusion of the gene encoding the exogenous polypeptide or protein with the phage gene encoding the coat protein, the final expressed protein will be presented on the surface of the phage as a fusion protein. And the polypeptide or protein encoded by the exogenous gene can remain the original spatial structure and biological activity. This group of phage introduced with various foreign genes is called a phage display library. Fix the molecule to be studied on the phage and use the phage to screen the phage display library. The phage carrying ligand that can specifically bind to the molecule is screened out, and the phage in the display library is isolated. So that the biological function of the foreign gene contained in the treated phage can be studied. The greatest feature of phage display is the unification of genotype and phenotype. At the same time, because of its simple operation, effective and easy to control, it is currently widely used in epitope analysis [53, 54], molecular interaction [55], and preparation of monoclonal antibodies. [56], drug screening [57], vaccine development [58], disease pathogenesis [59] and functional genomics [60] and other research fields.
When the phage display technology is applied to target screening, the phages fixed with the molecules to be studied are added to the phage library for screening. The foreign polypeptide of some phage can bind to the foreign polypeptide on the phage to be studied, and the phage that fails to bind will be discarded. After that, the bound phages are eluted with reagents, the eluted phages are collected to infect bacteria for amplification, and the obtained phages are repeated the above-mentioned screening process again. In this way, after 3–5 rounds of “adsorption-elution-amplification” circular screening, phages that specifically bind to the treated phage exogenous polypeptide can be found. After gene sequence analysis, the corresponding basic amino acid sequence is obtained according to the sequencing result, which is the amino acid sequence that can specifically bind to the target molecule. Finally, with the help of bioinformatics, analyze the proteins containing the above-mentioned amino acid sequences in the organism, and these proteins may be the target proteins of the molecules in the organism. Then perform experimental verification to verify the newly discovered target [2].
According to its display system, phage display technology can be divided into filamentous phage display system [61], λ phage display system [62], T4 phage display system [63], T7 phage display system [64]. The filamentous phages are single-stranded circular DNA viruses. The positions related to phage display mainly focus on pIII and pVIII capsid proteins, which are located at both ends of the phage particles and mostly fuse with exogenous gene sequences at the N-terminal or near N-terminal. However, the displayed molecule cannot be too large. If the displayed polypeptide is too large, it will affect the assembly of the coat proteins and make it lose its infectious power. It can fuse the exogenous gene sequence at the N-terminal, near N-terminal or the flexible joint region between N-terminal and C-terminal, and has no strict restriction on the size of the exogenous fragments displayed [65, 66]. The λ phage was found in 1951, it is a linear double-stranded DNA virus. The parts related to phage display are mainly concentrated in D and PV capsid proteins. The fusion of foreign protein and D protein will not interfere with the assembly of the λ phage, and the displayed proteins can be close to each other in space. The PV protein constitutes the tubular tail of the λ phage, and the C-terminus of the PV protein folding structure can be extended or replaced with foreign protein sequences. Since lambda phage is assembled in the host, it can display active macromolecular proteins (above 100 ku) and proteins that may be toxic to the host, and it is widely used [67, 68]. The T4 phage is also a double-stranded DNA virus. This display system relies on two non-essential capsid proteins, SOC and HOC. The exogenous protein or polypeptide is fused with the C-terminus of HOC or the N-terminus of SOC and displayed on the surface of phage. The T4 phage can be packaged in vitro to achieve specific site display. HOC protein and its fusion derivatives can be expressed in the host body, and then purified and added to the phage particles lacking HOC, which can still achieve the purpose of display. However, it is not commonly used because it adopts C-terminal fusion and has a small range of use [69]. The T7 phage is a linear double-stranded DNA virus, which is extremely destructive to host cells. In liquid culture, it only takes 1 to 2 h for the host bacteria to rupture from infection. It does not depend on the secretion mechanism of the host because T7 phage assembly in the cytoplasm, it is released by lysing bacteria. And it can survive harsh conditions such as high pH, high salt concentrations, and even in the presence of denaturants. The main display positions are the P10A and P10B capsid proteins. The functional capsid region of the phage can be composed of P10A, P10B alone or both in a certain proportion on the surface of the phage, which can accommodate a variety of variants of display proteins. Even if the insert fragments contain stop codons, it can also be displayed and expressed. In addition, the T7 phage surface can display various proteins of different molecular weights in low, medium and high copies. High copy number display is suitable for binding domains with low affinity, and low copy number display is suitable for high affinity binding domains. Therefore, the T7 phage display system is widely used [67, 68, 70] (The feature and limitations of the four phages are shown in Table 2).
| Types of phages | The positions related to phage display | Feature | Limitations |
| Filamentous phage | pIII and pVIII capsid proteins | Single, double, or triple display of different peptides on a single phage can be realized by site-specifically modification of peptides. | The displayed molecule cannot be too large. |
| λ phage | D and PV capsid proteins | It can display active macromolecular proteins (above 100 ku) and proteins that may be toxic to the host. | It is not easy to screen high affinity ligands. |
| T4 phage | SOC and HOC capsid proteins | It can be packaged in vitro to achieve specific site display and has large system capacity (above 35 ku). | It is not commonly used because it adopts C-terminal fusion and has a small range of use. |
| T7 phage | P10A and P10B capsid proteins | It can survive harsh conditions such as high pH, high salt concentrations, and even in the presence of denaturants. | T7 phage is extremely destructive to the host. |
| Even if the insert fragments contain stop codons, it can also be displayed and expressed. | |||
| It can display various proteins of different molecular weights in low, medium and high copies. |
The genotype and phenotype of the phage display technology are unified, and the gene information of the compound target can be directly obtained. According to the sequence information screened out, the possible target protein is obtained by combining the protein expressed by the sequence in the body. After further research, the mechanism of action of the compound can be revealed. Phage display technology is simple to operate and can save a lot of work and time compared with general sieve. However, it can only be applied to traditional Chinese medicine whose main active ingredient is protein. After improvements, such as biotinylation, it can also be applied to the discovery of TCM targets [71]. TCM whose active ingredient is not protein can also be used. Zhao et al. [72] screened the trichosanthin protein (TCS) using the pentadecapeptide library and the dodecapeptide library, and found that the specific binding polypeptide of TCS had homologous sequence with the phosphorylation site of protein kinase C (PKC). It shows that PKC may be a potential target protein of TCS. Pan screened the targets of San Shui Bai Hu Decoction to intervene in rheumatoid arthritis synovial cells from the phage random dodecapeptide library by using the whole cell subtraction screening idea [73]. The results screened out a specific short peptide “SGVYKVAYDWQH”, and the related proteins of this short peptide were Vaspin, MTMR14, BPAGI, DST, CAD, etc., which preliminarily indicated the target of Sanshui Baihu Decoction to interfere with rheumatoid arthritis synovial cells. Li et al. [74] used the T7 phage display to screen Salvia miltiorrhiza Bge. and Panax ginseng C. A. Meyer components for the treatment of lung cancer, and successfully screened out peptides with high affinity to Salviae miltiorrhizae Bge. and Panax ginseng C. A. Meyer compound components, indicating that it may be the target of Salviae miltiorrhizae Bge. and Panax ginseng C. A. Meyer for the treatment of lung cancer. Rodi et al. [71] synthesized biotinylated paclitaxel and then performed phage display, successfully identifying Bcl-2 as the target of paclitaxel in the body. However, due to the small size of the display library and the biotinylated compound is not stable enough, easy to decompose or has different properties from the original compound, which cannot support the experiment, or some compounds cannot even be biotinylated, so the application of phage display has certain limitations.
Drugs need to be combined with targets in the body to play a role. Bio-molecules have a specific structure, and only molecules that can bind to it can affect changes in the body. For example, only after the combination of enzymes and substrates can it play a catalytic role; only after the combination of antibodies and antigens can it play a therapeutic role; only after the combination of ligands and receptors can it play the role of up-regulation and down-regulation of body functions. And this kind of combination requires a certain affinity between the molecule and molecule, and the structure of the molecules can fit, and this combination is reversible. The affinity fishing is carried out according to this principle. The target molecule is fixed on the carrier by a certain method and exposed the active site. After passing protein mixtures such as cell lysate or tissue lysate through the carrier, the protein molecule that can affinity bind to the target molecule will be hooked onto the target molecule. Remove the unbound molecules, and then change the conditions to elute the bound protein molecules, and then molecules that can be bound by MS. Finally, the blocking method and other experiments are used to verify whether the drug molecule binds to the target protein and whether the drug molecule plays a role through the target protein.
Targets of active ingredients of TCM can be excavated by this method. The small molecule compound of the active ingredient of TCM can be fixed on different carriers by direct or indirect methods, and then the target protein can be screened out by “fishing”. However, there are few reports on this technical method, and it is still in the stage of development. Wulff et al. [75] used biotin labeling to attach (+) -avrainvillamide to the solid phase carrier, “fishing” the protein in the cell lysate. Based on protein mass spectrometry and other techniques, it was identified that a target of (+) -avrainvillamide in cells is nucleophosmin. Wang et al. [76] also used the method of biotin labeling to attach artemisinin to the solid phase carrier, and “fished” up to 124 suspected protein targets from the malaria parasite. Chen et al. [77] “fishing” the proteins in the brain that may interact with ginsenoside by the direct connection method. SDS-PAGE gel electrophoresis is used to separate the “fishing” proteins and identify them by MS. The periodic acid oxidation method improves the steric hindrance during intermolecular coupling, making it easy to “fish”. Finally, it was found that 14-3-3 protein is one of the targets of total ginsenosides in the brain. Wei used biotin labeling to “fish” the receptor for icariin to promote the maturation and mineralization of osteoblasts [78]. As a result, the biotinylated icariin was successfully synthesized and the biological activity remained unchanged. C-X-C chemokine receptor type 4 may be its receptor to promote the maturation and mineralization of osteoblasts. Zhou fixed the new tumor suppressor compound TCI04 on the CM5 chip, and used MDA-MB-231 human breast cancer cell lysate to “fish” the relevant target [79]. After identification by MS (Mass Spectrometry), it was finally found that Myoferlin protein may be the active target of TCI04 to inhibit tumor metastasis. The principle of affinity fishing technology is similar to that of HPLC (High Performance Liquid Chromatography), but affinity fishing technology is based on the affinity between molecules and molecular structure to screen biological targets, and affinity fishing technology also needs to be fixed on a carrier.
Affinity fishing technology also has certain problems. For example, most molecules currently need to be labeled or even biotinylated. After these pre-treatments, the properties of the molecules may change, so the protein targets fished may be different from those of the original molecules. Moreover, this technology cannot achieve high-throughput screening. If a large number of protein targets are to be screened, it will take a long time. However, the above problems can be solved, and this technology can be combined with microarray chip technology to achieve the purpose of high-throughput screening. Biacore is a biosensor based on surface plasmon resonance (SPR) technology, which can detect biomolecules in real time, especially to finely characterize the interaction between targets and small molecules, and provide kinetic and thermodynamic data on drug target interactions [78]. This technology does not require the use of fluorescent labels and isotope labels, so it can guarantee the accuracy of affinity fishing to the greatest extent. As an in vitro target prediction technology, affinity fishing technology is simple to operate. It does not require too much experimental equipment and too much expense, and it is feasible for preliminary prediction of protein targets (Fig. 4).
 Fig. 4.
Fig. 4.The process of affinity fishing.
The drug affinity responsive target stability (DARTS) was first proposed by Lomenick [80] in 2009. Its basic concept is that after the drug binds to the target protein, the target protein may become protease resistant. Then use protease to hydrolyze the unbound protein, and the unhydrolyzed protein is the target protein, so it can be used to screen the target of drug molecules. Compared with affinity fishing, DARTS operation is simpler, no compound modification is required, and multiple compounds can be screened at the same time.
Compared with affinity angling, DARTS is more simple to operate, does not need to modify compounds, and can complete the screening of multiple compounds at the same time. The general procedure of the DARTS experiment is to add a buffer solution to the cultured cells, and then add a lysis solution to lyse the cells for a period of time (such as M-PER lysis buffer [81]) for about 10 min. A certain protease inhibitor is added to the lysis solution to inhibit the activity of protease in the cell. After full lysis, the supernatant was collected by centrifugation, and the enzyme-protein reaction buffer was added in proportion. After full lysis, centrifuge to take the supernatant, and the enzyme protein reaction buffer was added in proportion. The supernatant was divided into two parts, one was added with DMSO as a control group, and the other was added with drug molecules for co-incubation. After the small drug molecules are combined with the cell lysate for a period of time, a protease is added for enzymatic hydrolysis for a period of time, and then the reaction is terminated with a protease inhibitor or high temperature. Finally, the binding protein was detected and identified by protein research techniques, such as SDS-PAGE and Western blotting, and the protein target of the drug was identified by coupling with MS.
The key point in this process is the choice of protease. There are currently three types of proteases reported for DARTS technology, namely subtilisin, thermolysin, and streptomycin. The subtilisin is a series of serine protease mixtures first obtained from Bacillus subtilis. Under certain conditions, it catalyzes the hydrolysis of proteins rapidly by attacking the peptide bond by the serine residues in the active site. It has strong proteolytic activity, but it is limited by the need to operate under alkaline conditions [81]. The thermolysin is an extracellular high-temperature-resistant metallopeptidase produced by thermophilic bacteria with low cleavage activity. The enzyme can only effectively cleave protein polypeptide chains in the unfolded state. In the DARTS experiment, the thermolysin can hydrolyze a variety of cellular proteins under standard reaction conditions, but its stability is greatly improved after small molecules are combined with non-target proteins, which is not conducive to degradation [82]. The streptomyces protease is an extracellular protease mixture of serinase and acid protease isolated from the streptomyces griseus. It can specifically cleave the peptide bonds on the carboxyl side of aspartic acid and glutamic acid in both folded and unfolded protein and polypeptide chains [83], so the streptomyces protease is the optimal recommended protease (Fig. 5).
 Fig. 5.
Fig. 5.The process of DARTS.
The characteristics of DARTS technology allow it to have great advantages in the
discovery of drug targets. The binding strength of proteins to small drug
molecules depends on the affinity of the two. Wang used DARTS technology to
search for binding proteins of the new structure anti-tumor lead compound
Rasfonin, and found that 11 proteins are closely related to tumors or related
signal pathways [84]. It can be used as a clue for in-depth study of Rasfonin
binding protein, and the result is better than the method using affinity
chromatography. Chin et al. [85] found that tricarboxylic acid cycle
intermediate
The DARTS technology has rarely been reported on the discovery of Chinese medicine targets, but the advantages of this technology can support its screening in compound prescriptions. However, before application, it is necessary to clarify the active ingredients in the compound. Therefore, the DARTS technology has certain advantages and effects in the research of TCM compound prescriptions. With the gradual improvement of DARTS technology, this technology will surely shine in the identification of traditional Chinese medicine target proteins.
Drugs and targets can be stably combined to affect their functions in order to achieve the purpose of treatment, but there are two common bottlenecks that have been plaguing the development of new drugs. One is how to make drugs correctly recognize target proteins; the other is whether they can bind to targets after recognition. Targeted detection of drugs in cells is often difficult because drug binding cannot be measured directly within cells. At present, the efficacy of drugs is indirectly monitored by studying the reactions of downstream cells or cell products, but many drugs have been found in subsequent studies that they cannot be combined with the target [88]. Therefore, in order to be able to monitor the binding of intracellular drugs to targets, scientists at the Karolinska Research Center developed a technology called cellular thermal shift assay (CETSA) in 2013 to monitor the binding of drugs to target proteins in cells and tissue samples [89].
Proteins perform their biological functions by folding into specific
three-dimensional structures, and active proteins usually fold well at room
temperature. When proteins are exposed to higher temperatures, they will expand
and lose their three-dimensional structure. At this time, proteins will aggregate
and precipitate [90, 91], which is called unfolding event. It is similar to DARTS
technology, which is based on the principle that after the ligand binds to the
target protein, it can make the protein more stable. However, there are some
differences between the CETSA and DARTS. The CETSA is based on the fact that
after the ligand binds to the target protein, it will stabilize the protein and
reduce the free energy
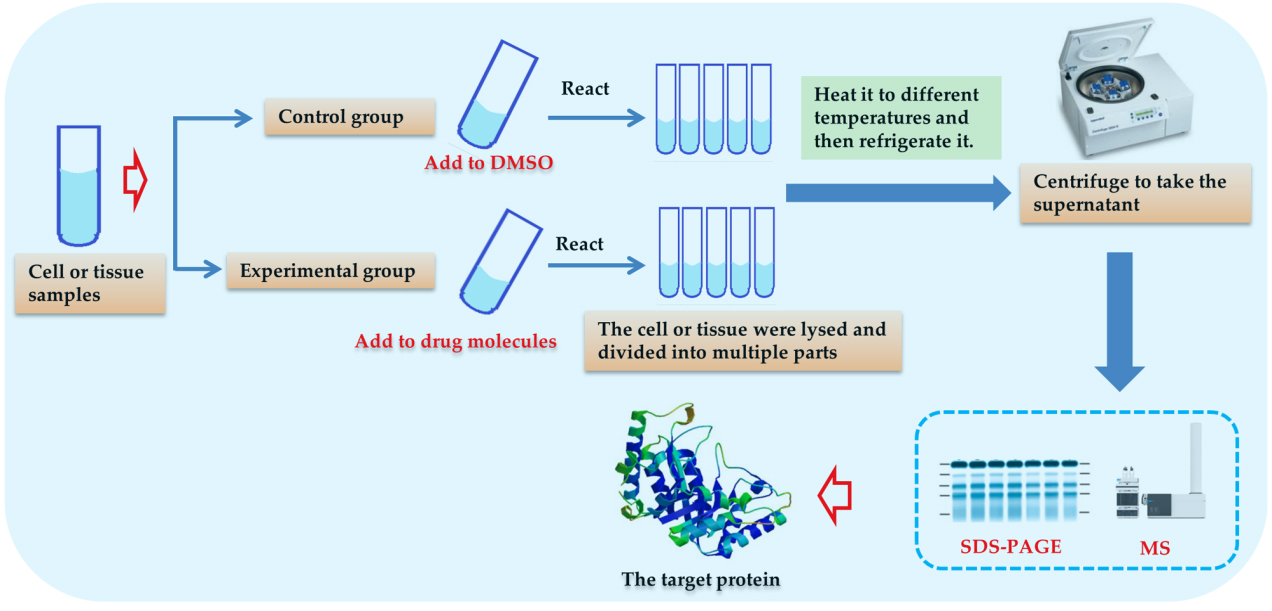 Fig. 6.
Fig. 6.The process of CETSA technology.
 Fig. 7.
Fig. 7.The CETSA curve and the ITDRFCETSA.
There are currently some studies using the CETSA technology to discover and verify targets, all of which have achieved ideal experimental results. Atikul et al. used the CETSA technology to verify the binding of capsaicin to the cell cycle-related enzyme tNOX, and found that capsaicin can bind to tNOX in T24 bladder cancer cells. This combination inhibits the production of NAD+ and down-regulates tNOX, thereby inhibiting sirtuin-1 (SIRT1) deacetylase activity [93]. Ultimately, decreased SIRT1 deacetylase activity will enhance the acetylation of c-Myc and p53, leading to cell cycle arrest, thereby inhibiting the growth of cancer cells. Dziekan et al. [94] used the combined CETSA-MS technique with drugs that bind to Plasmodium falciparum lysates and fully infected red blood cells. A common protein target of quinine and mefloquine against Plasmodium falciparum was found to be purine nucleoside phosphorylase. Rheumatoid arthritis is a chronic joint synovial inflammatory disease with unknown etiology. Radix Litseae Cubebae. is the dried root and rhizome of Litsea cubeba (Lour.) Pers., and is one of the TCMs. Previous study predicted the key targets of RadixLitseae Cubebae. for the treatment of rheumatoid arthritis using molecular docking technology, and the predicted targets were verified by CETSA technology [95]. The results showed that LC36, the main active component of RadixLitseae Cubebae., could bind to MEKl and cathepsin K in cells to play a role in the treatment of rheumatoid arthritis, indicating that the targets of LC36 are MEK1 and cathepsin K. RAS oncogenes are frequently mutated in human cancers. Through a large number of experimental studies, H. Yurugi et al. [96] found that flavaglines, the main active component of Aglaia species used in TCM, can be very good for inhibiting the activation and stability of RAS (RAS is an oncogene) through prohibitins-1 protein. At the same time, CETSA was used to verify the direct targets of flavaglines, and the experiments identified prohibitins-1 as the direct targets of flavaglines.
This technology does not require structural modification or labeling of molecules, and avoids changes to molecular properties. The CETSA technology can detect whether the ligand binds to the target by detecting the change in the free energy of the system after the ligand binds to the protein target. Therefore, this technology is the most suitable for verifying whether the ligand binds to the target in vivo. Some drugs can bind to the target during the research phase, but later studies have found that they cannot bind to the target in the body, so they cannot exert their efficacy. Using this technology can verify the authenticity of the target binding. Because it can directly monitor the binding of drugs and targets in the body, this technology has great advantages for the discovery of TCM targets. Various methods can be used to predict the target of traditional Chinese medicine in the early stage of the research. Finally, CETSA technology is used to verify the accuracy and authenticity of the target. As the technology is still in the development stage, the advantages and disadvantages are still unclear. But it is undeniable that this technology will have great application in target discovery and verification.
At present, there are some other technologies that can be considered for target research, such as molecular imprinting technology and surface plasmon resonance technology, etc., which will be briefly introduced in this section.
The concept of “molecular imprinting” was first proposed by Dickey [97] in 1949 and then developed rapidly. The principle is that template molecules are combined with functional monomers, and then polymerization under the action of cross-linking agents gives polymers with large pores and meshes. After that, the template molecules in the medium were eluted by physical or chemical methods to obtain the polymer with the spatial structure and binding site of the template molecules. Functional monomers and template molecules are combined through hydrogen bonds, electrostatic interactions, hydrophobic interactions and other non-covalent interactions, so that these functional molecules complement each other with the structure of template molecules [98, 99]. At present, this technology is mainly used in the fields of separation, simulation of antibody and receptor, catalyst and artificial enzyme, etc. Through the analysis of its principle, we found that this technology can also be applied to the research of the target of TCM. A polymer of a compound whose target is known is constructed, and then the polymer is reacted with the ingredient groups of traditional Chinese medicine. After the bound chemical components are eluted, the chemical components similar to the known compound structure can be obtained, and finally the validity of the compounds is verified.
Surface plasmon resonance (SPR) is a physical optical phenomenon. When total reflection occurs when a beam of mid face monochromatically polarized light is illuminated over a certain range of angles onto a film of metallic silver or gold plated on a glass surface, the light is both coupled and enters the metallic film when the wave vector of incident light matches the oscillation frequency of surface electrons (called plasmons) inside the metallic film, causing the electrons to resonate, which is known as SPR. At this point the energy provided by the light causes the metal film surface electrons to resonate and the electrons absorb that energy to minimize the intensity of the light being reflected, and the angle of incident light at which this minimization occurs is called the SPR angle. The SPR angle varies with the refractive index of the metal surface, which in turn is proportional to the mass of biomolecule bound to the metal surface [100]. The analysis process of the instrument based on this technology is that the target molecule is fixed on the surface of the sensor, and the protein target that can bind to the molecule causes the SPR Angle to change through the protein solution. Therefore, the interaction between two molecules can be monitored, and it can also be used for target screening.
Currently, the TCMs have aroused increasing interests of pharmaceutic scientists, and the TCMs have been considered as a precious resource for finding novel candidate drugs [101, 102]. This paper briefly described several currently commonly used target research methods, such as computer aided drug design, network pharmacology, phage display, affinity fishing, drug affinity responsive target stability and cellular thermal shift assay. Among them, each technique has its own characteristics, and some techniques can be applied for target discovery, while others can be carried out to target verification. Each method can be used singly or in combination to ensure the accuracy of target points. Each method can be used alone or in combination with multiple methods to ensure the accuracy of the target. For example, after using computer aided drug design to screen out drug targets, cellular thermal shift assay is used for in vivo experimental verification and microscale thermophoresis technology is used for in vitro experimental verification, so that the discovered target is the most authentic target. With the advent of the era of big data, the discovery of drug targets is not limited to biotechnology. Only by interdisciplinary and multi-technology combination can the efficiency and accuracy of target research be guaranteed (The specific advantages and disadvantages are shown in Table 3).
| Technology or strategy Design | Advantages | Disadvantages | Application direction | Time |
| Computer Aided Drug (CADD) | 1. The workload is small and the speed is fast. | It is only a prediction, not a definite target, and needs to be validated | At the beginning of the study, prediction and sear- | In the early 1980s |
| 2. Strong predictive ability. | experimentally. | ch for the target. | ||
| Network pharmacology | 1. The characteristics of “Multi - component, multi - target and multi – pathway” are suitable for target research of traditional Chinese medicine. | The target is not real and needs to be verified experimentally. | At the beginning of the study, prediction and search for the target. | In 2007 |
| 2. Able to analyze the function of genes or targets. | ||||
| Phage display | 1. The unification of genotype and phenotype. | The display library capacity is small | Target search or experi- | In 1985 |
| 2. It is simple operation, effective and easy to control. | and needs to be biotinized. The biotinized compounds are not stable or have different properties from the original compounds. | mental verification. | ||
| The Affinity fishing | 1. It can detect biomolecules in real time and fine-characterize the interactions between the target and small molecules | It needs labele or biotinization. | Preliminary prediction of protein targets. | In 1983 |
| 2. It can provide kinetic and thermodynamic data on drug target interactions | High throughput screening is not possible. | |||
| The Drug Affinity Re- | 1. The operation process is simple. | High requirements for experimental | Screening of protein tar- | In 2009 |
| sponsive Target Stability (DARTS) | 2. No modification is required for the compound. | operation. | gets and verification of targets. | |
| 3. Multiple compounds can be screened simultaneously. | ||||
| Cellular thermal shift assay (CETSA) | 1. It can monitor the binding of drugs to targets in cells and tissues. | Currently unclear. | In vivo validation of protein targets. | In 2013 |
| 2. No modification is required for the compound. |
At present, the difficulty of target research is not in the discovery of the target. It is easy to find the target by a variety of technical means. The key points are whether the drug target that has been discovered can be combined with the drug in the body and exert its efficacy. Cellular thermal shift assay is a newly developed technology, which can study the combination of drug molecules and targets in vivo. However, the advantages and disadvantages of this technology are not clear, and it is not enough to rely solely on this technology. The multi-component, multi-target, and multi-pathway characteristics of TCM make it effective in the treatment of many diseases. However, it is precisely because of its characteristics that it is difficult to study TCM targets. All of the techniques described above can be applied to the research of TCM targets, but because of the complexity of TCM components and targets, the above techniques have some sidedness and limitations, and to correctly and effectively study the targets of TCMs, a combination of multiple techniques is required to achieve the goal. Although these technologies all increase the efficiency and accuracy of target screening relative to universal screening, the combined use of multiple technologies can also make the effort prohibitive. All of these techniques can improve the efficiency and accuracy of target screening compared to general screening, but the combined use of multiple techniques can also cause a large workload. Therefore, future research directions can focus on the development of simple and effective new technologies (similar to CETSA technology) or combine and optimize existing technologies to provide convenience and support for drug target research.
All authors contributed substantially to the preparation of this review. FL has concetulize the review, drafted and edited the review. DW, RLL, LYH have drafted and edited the review. LA, CJW has revised and confirmed the review. All authors discussed and confirmed the final manuscript.
Not applicable.
Not applicable.
This research was supported by the Project of Administration of Traditional Chinese Medicine of Sichuan Province of China (No. 2020HJZX001), the Sichuan Science and Technology Program (grant number 2020YFS0523 and 2020YFS0525), Xinglin Scholar Discipline Promotion Talent Program of Chengdu University of Traditional Chinese Medicine (No. BSH2018006),the Project of Sichuan Provincial Administration of Traditional Chinese Medicine (2021MS017) and the Project of Science and Technology Department of Sichuan Province (2021YJ0112).
The authors declare no conflict of interest.
TCM, traditional Chinese medicine; CADD, computer aided drug design; NBP, n-Butylphthalide; BMECs, brain microvascular endothelial cells; OGD, oxygen glucose deprivation; VSMC, vascular smooth muscle cells; PDGF, platelet-derived growth factor; ox-LDL, oxidized low density lipoprotein; DARTS, drug affinity responsive target stability; CETSA, Cellular thermal shift assay; MST, Microscale thermophoresis.
