† These authors contributed equally.
Background: Myocardial Infarction (MI) is a cardiovascular disease with
a high morbidity and mortality rate. While MI is currently treated with
pharmaceuticals, there is a need for new treatment options: compound Chinese
medicines may have unique advantages for the treatment of MI. Methods: A
combination of network pharmacology and experimental verification is used to
identify the ingredients and mechanism of Compound Longmaining (CLMN) for
treating MI. Network pharmacology combined with the gene expression omnibus (GEO)
chip method is used to analyze the primary pathway of CLMN for treating MI, and
then molecular docking is used to verify the affinity of key target proteins in
the primary pathway that bind to active molecules. The major active compounds of
CLMN are screened using the docking score results. The CIBERSORT algorithm is
used to evaluate immune cell infiltration in MI, and high performance liquid
chromatography (HPLC) is used to control the quality of the components. Finally,
a mouse model is established to verify the molecular mechanism of CLMN for
treating MI using hematoxlyn eosin (HE) staining and immunohistochemistry.
Results: By utilizing network pharmacology combined with molecular
docking, the mechanism of action of CLMN for the treatment of MI was found to
possibly be related to the ingredients of puerarin, daidzein, ferulic
acid, chrysin, and galangin. These molecules regulate the NF-Kappa B signaling
pathway and the expression of RELA, IKBKB, NKBIA, and
other targets. The CIBERSORT algorithm and ggplot2 package analysis were used to
distinguish the immune cells, such as neutrophils, macrophages, and T cells, that
play a key role in the development of MI. HPLC controlled the quality of the
screened medicinal ingredients. An immunohistochemical analysis showed that the
TNF-
Cardiovascular disease is the leading cause of death among urban and rural residents in China, a trend that has increased recently [1]. In 2014, there were approximately 290 million patients with cardiovascular disease, which included 2.5 million myocardial infarction cases. In the United States, there are approximately 1.5 million cases of MI annually with a yearly incidence rate of approximately 600 cases per 100,000 people [2]. Although the prognosis of MI has been greatly improved, it is still the primary cause of morbidity and mortality in the world [3]. Therefore, it is important to continue to develop new therapeutics based upon the use of novel strategies to understand the mechanisms involved in MI. Following MI, there is cardiomyocyte necrosis, the result of ischemia that causes an inflammatory response. Oxidative stress mediated by inflammatory factors damages vascular endothelial cells, and leads to plaque rupture and hemorrhage [4]. These triggering events cause serious clinical manifestations.
Traditional Chinese Medicines (TCMs) have the potential advantage of low toxicity and broad spectrum activity to prevent and treat a wide variety of diseases including MI. CLMN is composed of the following natural ingredients: Pueraria lobata, Dioscoreae nipponica Makino, Ligusticum wallichii, and propolis. The active molecules identified in CLMN are flavonoids, puerarin, alkaloids, lactones, phenols, organic acids, steroidal saponins, flavonoids and terpenes [5]. The CLMN is an in-hospital preparation developed by Professor Tao Genyu of the affiliated Hospital of the Shaanxi University of Chinese Medicine. Since previous studies have confirmed that the CLMN decoction has a significant therapeutic effect on MI [6], we used network pharmacology combined with a gene expression omnibus (GEO) chip analysis and molecular docking to explore which components, targets, and pathways that are regulated by CLMN.
Network pharmacology can provide insight into “component-target-pathways” of drugs and help predict the correlation between drugs and diseases [7]. The microarray analysis has been widely used to identify differentially expressed genes (DEGs) and functional pathways involved in disease progression. Finally, molecular docking is a technique used to visualize drug interactions with target proteins [8]. This study explores the possible mechanism of CLMN for the treatment of myocardial infarction and uses the CIBERSORT algorithm to evaluate the immune cell infiltration in MI from the GEO database. This study also uses the high performance liquid chromatography (HPLC) method to establish the quality control of CLMN and conduct an animal experiment verification to provide a more reliable basis for clinical application. The flow chart of the study is shown in Fig. 1.
 Fig. 1.
Fig. 1.A detailed flow chart of the study.
The Traditional Chinese Medicine Systems Pharmacology (TCMSP,
http://lsp.nwu.edu.cn/tcmsp.php) database [9] and scientific literature allowed
us to identify the chemical constituents of CLMN and the targets of four TCMs:
P. lobata, D.nipponica Makino, L.
wallichii, and propolis. The oral Bioavailability ((OB)
In order to obtain gene data related to MI, we searched GeneCards
(https://www.genecards.org/) [14], Comparative Toxicogenomics Database
(http://ctdbase.org/http://ctdbase.org/), and the DisGeNET
(https://www.disgenet.org/) public database using the keyword “Myocardial
Infarction”. The GSE61145 (GPL6106) and GSE60993 (GPL6884) gene expression
datasets were downloaded from the GEO (https://www.ncbi.nlm.nlh.gov/geo/)
database [15], and 10 normal samples and seven preoperative MI samples in
GSE61145 were chosen. In addition, seven normal samples and 17 MI samples in
GSE60993 were collected from the peripheral blood of the subjects. Using the R
language “limma” package [16] on the analyses and the expression of genes after
normalization, screening disease DEGs, and map volcano, the screening conditions
were determined to be a p-value
The overlapped genes were deleted from the disease genes and component target genes obtained from the multiple databases. The genes that overlapped between the datasets were obtained using an online Venn analysis tool (https://bioinfogp.cnb.csic.es/tools/venny), and the common genes were screened by mapping.
The target genes of the CLMN components were introduced into the STRING (https://string-db.org/) database [18], and the research species was set to “Homo sapiens”. In the setting, the highest confidence level was set to 0.9, the free target genes were hidden, and the TSV format file of the relationship diagram was derived after obtaining the target-protein interaction relationship. We imported the Tab Separated Values file into Cytoscape3.7.2 (Donnelly Centre for Cellular and Biomolecular Research, Toronto, North America, Canada) [19] and adjusted the color of the nodes according to the degree value, in order to more intuitively show the important CLMN targets.
The target genes of the active components of CLMN were imported into Cytoscape 3.7.2. The node representing the drug active component was set to “source node”, the node representing the disease target gene was set to “target node”, and the network attribute was set to “interaction type” to draw the “drug active ingredient-core target” network.
The core target genes of drug-disease were transformed from Gene Symbol to
entrezID, using R language, and the data was processed using “BiocManager”. We
used the R package clusterProfiler package [20] to perform the GO and KEGG
enrichment analysis on the shared targets, The GO and KEGG enrichment analysis
were obtained under the condition of p
The protein structure of the key target was downloaded from the Protein Data Bank (PDB, https://www.rcsb.org/) database [21], and we download the sdf file for the ligand (active component and positive drug corresponding to the target) from the Pubchem database. The LibDock tool of Discovery Studio 4.0 software (Neo Trident Technology LTD, Beijing, China) was used to dock the key target protein with its corresponding active component and positive drug, and the score was analyzed.
Two microarray data sets, GSE61145 and GSE60993, were downloaded from the GEO
database to screen different infiltrated immune cells, and the CIBERSORT
algorithm [22] was used to evaluate the immune cell infiltration in MI. CIBERSORT
is an analytical tool that represents the cellular composition of complex tissues
based on pretreated gene expression profiles. This default signature matrix of
100 permutations was used in this algorithm, only data with p values
The CLMN included four types of medicinal materials, namely P. lobata, D. nipponica Makino, L. wallichii, propolis. The P. lobata, D. nipponica Makino, L. wallichii decoction pieces were purchased from the Baoji Xiangyuan Traditional Chinese Medicine Decoction Pieces Co., LTD., and the propolis was purchased from the Natural Son Peak Products Co., LTD., Xianyang, Shaanxi Province. The CLMN decoction was prepared as described previously. A total of 18 g of P. lobata, 12 g of D. nipponica Makino, 12 g of L. wallichii, and 6 g of propolis was added to14 volumes of water, heated and then boiled twice. The filtrate was consisted of concentrates of 0.32 g the crude drug/mL [24].
By utilizing the network pharmacology, molecular docking technology and a literature review, the effective and high content of the pharmacodynamic components in the CLMN were determined, and HPLC was used to control the quality of the effective components.
Male Balbc mice weighing 22–28 g and 10–14 weeks old were purchased fromthe Chengdu Dashuo Experimental Animal Co., Ltd., Chengdu, China. The experimental animal license number was SYXK (chuan) 2020-030. The animals were maintained under the standard laboratory temperature and humidity conditions and had free access to food and water throughout the study. This experiment was approved by the Animal Ethics Committee of the Shaanxi University of Chinese Medicine.
Sixty mice were randomly divided into six groups: a sham operation group, a positive control: a Compound Danshen Dropping Pills (CDDP) group, an MI model group, and low-dose, medium-dose and high-dose CLMN groups. The mice in the sham operation group and the MI model group were given normal saline. The mice in the CDDP group were given 9.78 g/kg/d, and the mice in the CLMN groups were given the CLMN decoction at 3.26 g/kg/d (low dose), 6.52 g/kg/d (medium dose) and 13.04 g/kg/d (high dose) for seven days.
The MI model was established as described previously. Briefly, we opened the pericardium and located the left anterior descending coronary artery and inserted a needle. Then the coronary artery of the left anterior descending branch of the left atrium was ligated using a 8/0 monofilament polypropylene suture. This resulted in permanent ischemia of the artery below the ligation line. For the sham group, the needle was inserted into the left anterior descending branch of the coronary artery without ligation after thoracotomy [25, 26].
Following MI, the mice were anesthetized and the fresh heart tissue was quickly removed and frozen in liquid nitrogen for the pathological examination. The myocardial tissue was then fixed in 4% paraformaldehyde and embedded in paraffin. Thereafter, the tissue was cut into 5 um thick sections and stained using hematoxylin-eosin (HE), and the histopathological examination was performed under a light microscope (OLUMPUS Japan) at 200 magnification.
The myocardial tissue was cut into 5 um slices, placed in xylene, and hydrated
with an ethanol gradient (100%, 95%, 80%, 70%, and pure water). After antigen
repair, the slices were incubated in H
Data was analyzed using SPSS Statistics 26
software (IBM Corp., Chicago, IL, USA), and the results are expressed as means
In this study, 37 active compounds were collected from the TCMSP and PubChem databases, including nine active compounds in P. lobata targeting 506 proteins, four active compounds in D. nipponica Makino targeting 109 proteins, 12 active compounds in L. wallichii targeted 443 proteins, and 12 active compounds in propolis targeting 1219 proteins. After removing the same target of four herbs, a total of 572 target proteins of CLMN were obtained.
Using the Gene Cards, CTD, and DisGeNET databases in combination, a total of 32,902 MI-related targets were obtained after the deletion of duplicate and invalid targets. A total of 1764 and 1079 significant difference genes were obtained by analyzing the GSE61145 and GSE60993 chips, respectively, and the DEGs volcanic map was drawn. As shown in Fig. 2. We examined the expression of DEGs from both datasets. Genes that were highly or lowly expressed in both datasets were retained as DEGs, resulting in 631 DEGs.
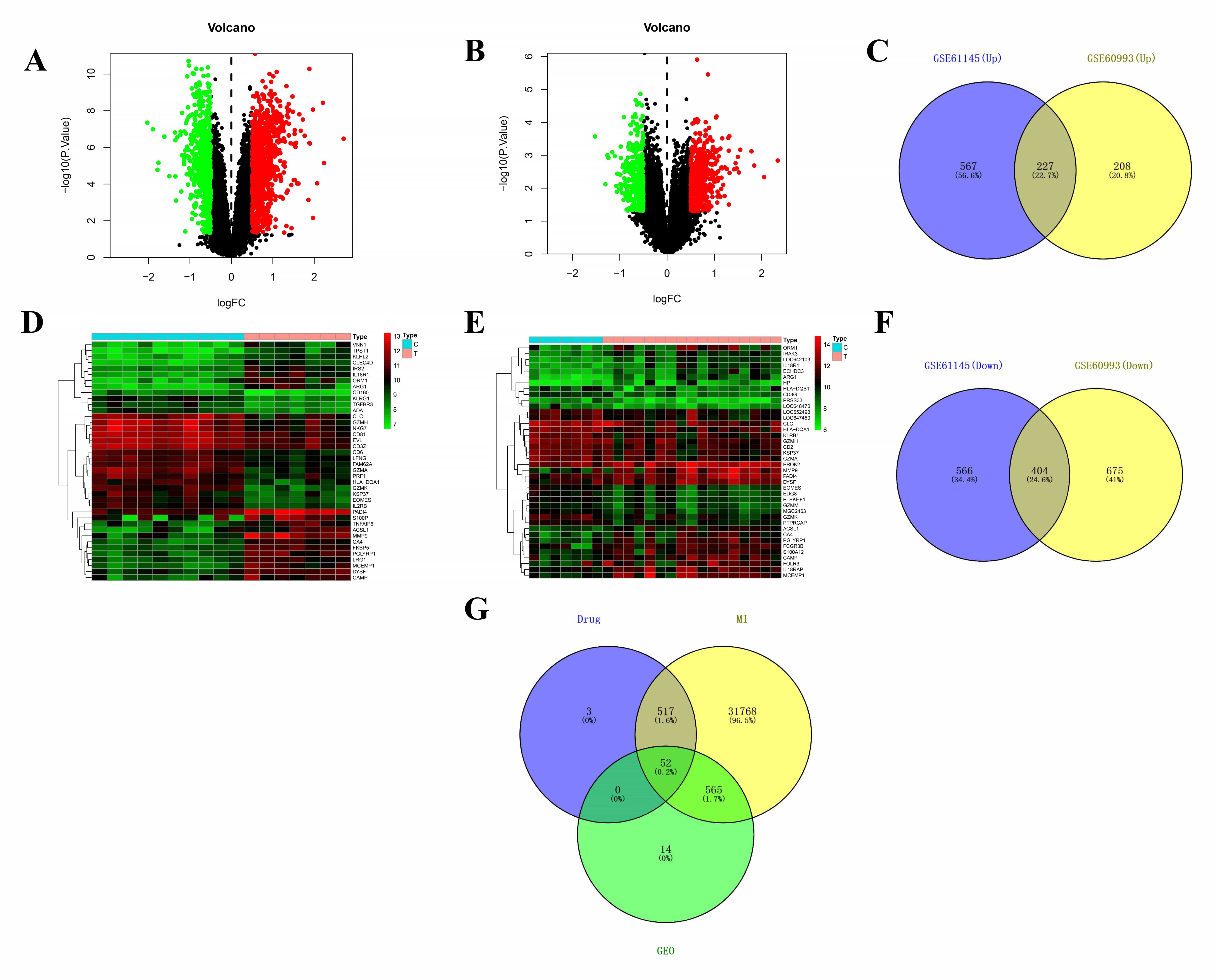 Fig. 2.
Fig. 2.Screening of core targets. (A) DEGs volcanic map of the GSE61145 chip. (B) DEGs volcanic map of the GSE60993 chip. (C) GSE61145 chip and GSE60993 chip up-regulate intersection genes. (D) DEGs heat map of GSE61145 chip. (E) DEGs heat map of GSE60993 chip. (F) GSE61145 chip and GSE60993 chip down-regulate intersection genes. (G) CLMN active ingredient target and disease target Venn diagram. In the volcano map, the down-regulated genes in the normal group are represented by green dots, while those up-regulated in the experimental group are represented by red dots.
There were 572 compositional targets from the TCMSP and Swiss Target Prediction databases, 32,902 disease targets from the GeneCards, CTD and DisGeNET databases, and 631 DEGs from the GEO database that were imported into an online Venn analysis tool. A total of 52 drug-disease interaction targets were obtained. The results are shown in Fig. 2C.
The 52 targets described above were imported into the STRING database and were
viewed in the network diagram “Analysis” of the database. The PPI network
consisted of 52 nodes and 37 edges. The average node degree was 1.42, and the
average clustering coefficient was a p-value
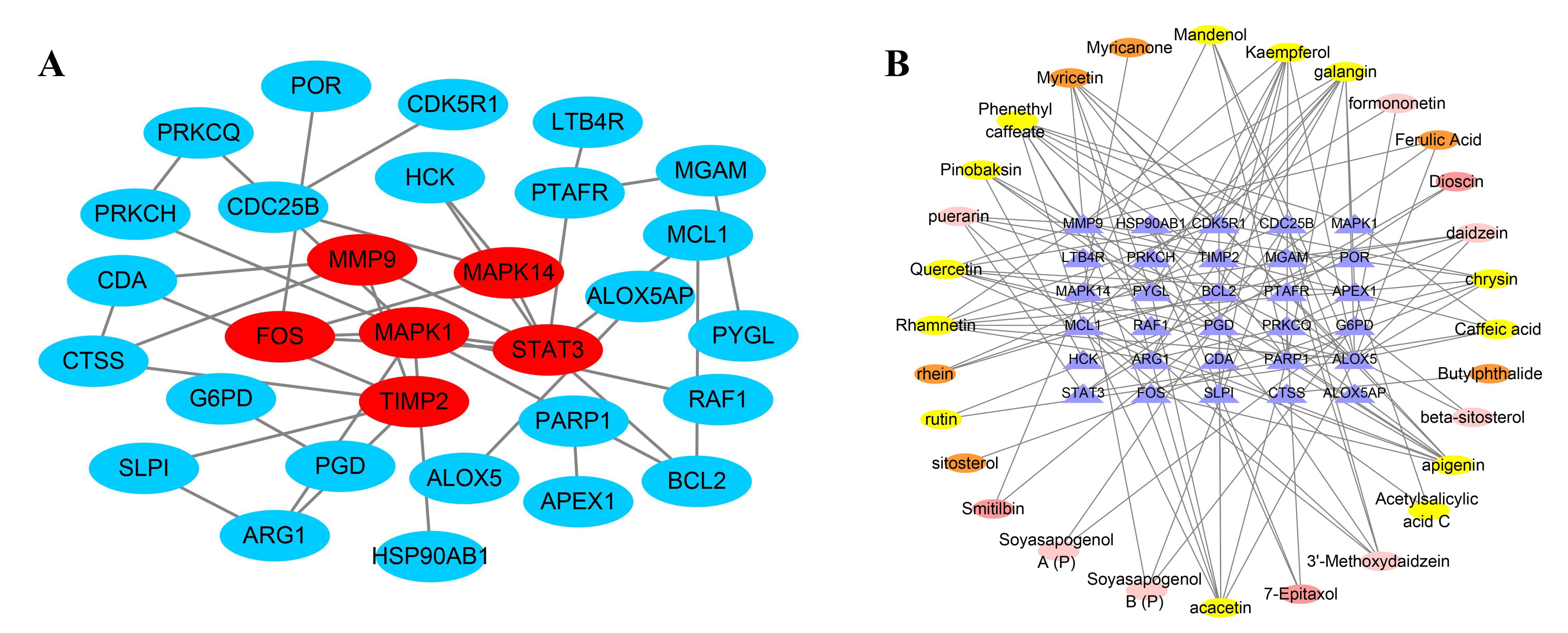 Fig. 3.
Fig. 3.CLMN-MI core target network diagram. (A) CLMN-MI core target PPI network diagram. (B) CLMN active component-core target interaction network diagram.
Using the “merge” tool within the Cytoscape3.7.2 software (Donnelly Centre for Cellular and Biomolecular Research, Toronto, North America, Canada), a network diagram for the 37 drug active components and 52 MI targets was constructed (Fig. 3B). Among them, the ovals of the different colors were used to represent each active ingredient from different TCMs, and the triangle represents the disease targets. The network consists of 159 nodes and 125 edges. From the graph, it is apparent that a single active component corresponds to multiple targets, and one target can also correspond to multiple active components. Taken together, CLMN therapy for MI has the characteristics of possessing multi-components and multi-targets. Using the Network Analyzer analysis tool, the top components were galangin, apigenin, phenethyl caffeate, and acacetin, and the top targets were ALOX5, MMP9, CDK5R1, ARG1 and BCL2. This result suggested that all may play an important role in the mechanism of CLMN for the treatment of MI.
According to a p
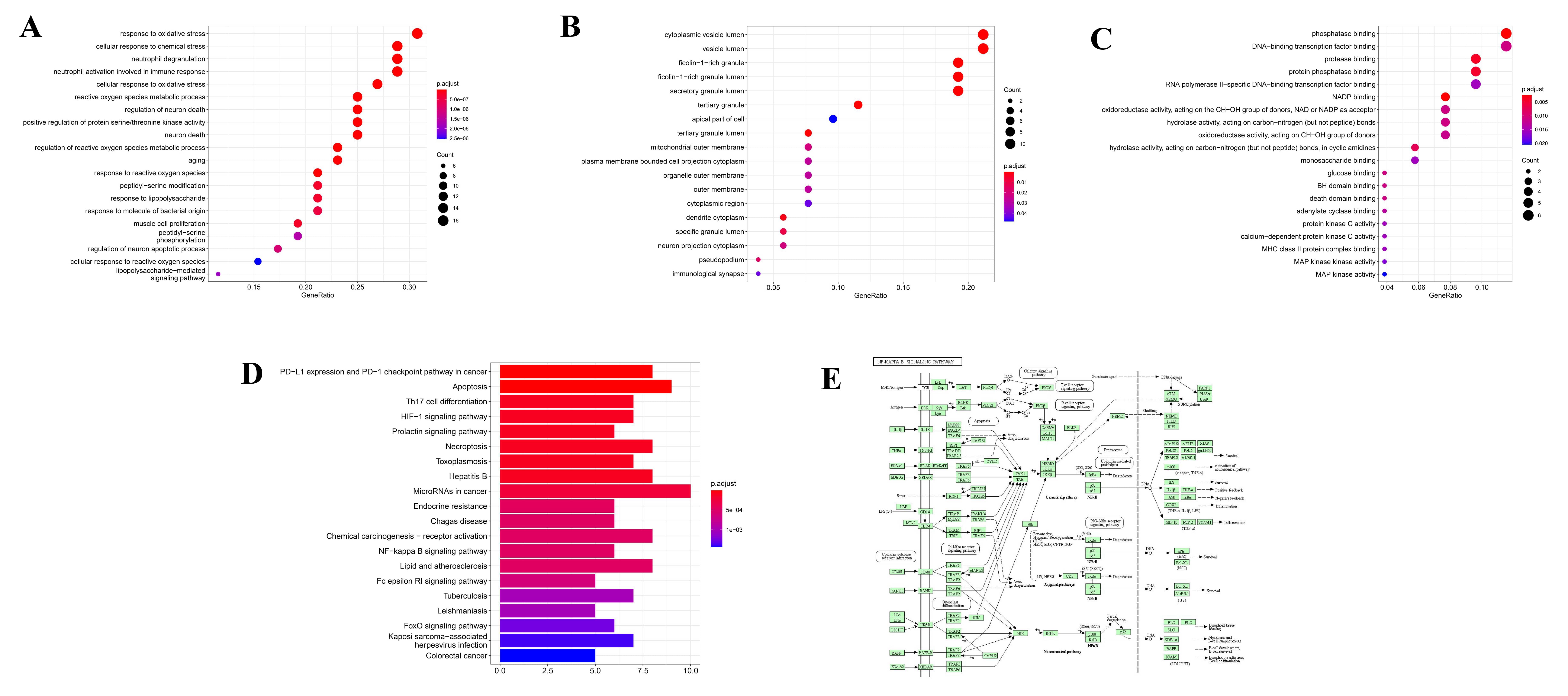 Fig. 4.
Fig. 4.GO analysis and KEGG pathway diagram of CLMN in the treatment of MI. (A–C) GO analysis bubble chart of CLMN treatment of MI. (D) KEGG pathway analysis histogram of CLMN treatment of MI. (E) NF-Kappa B signal pathway diagram.
The docking results are shown in Table 1, and the interactions between them are shown in Fig. 5. In order to further illustrate the binding activity between the target protein and its corresponding compound, in this study, the key targets RELA, IKBKB, NFKBIA, and their corresponding compounds in the NF-Kappa B pathway were selected. Discovery Studio 4.0 software (Neo Trident Technology LTD, Beijing, China) was used for positive drug verification experiments and molecular docking. The higher the binding activity, the higher the score. The docking scores of the target protein and its corresponding small molecule compounds were compared with those of the positive control. The results showed that the target RELA had good binding activity with puerarin, ferulic acid, and daidzein. IKBKB had good binding activity with chrysin and galangin, and NFKBIA had good binding activity with puerarin. In addition, the conformation of the combination was stable, indicating that these may be the key components and targets of CLMN in the treatment of MI.
| Key target | Small molecule ligand | Docking score | Positive for drugs | Docking score |
| RELA | ferulic acid | 101.9897 | SC-236 | 105.787 |
| daidzein | 98.4975 | |||
| puerarin | 108.584 | |||
| IKBKA | chrysin | 102.312 | Auranofin | 99.9923 |
| galangin | 98.5631 | |||
| NFKBIA | puerarin | 98.3206 | Astaxanthin | 101.948 |
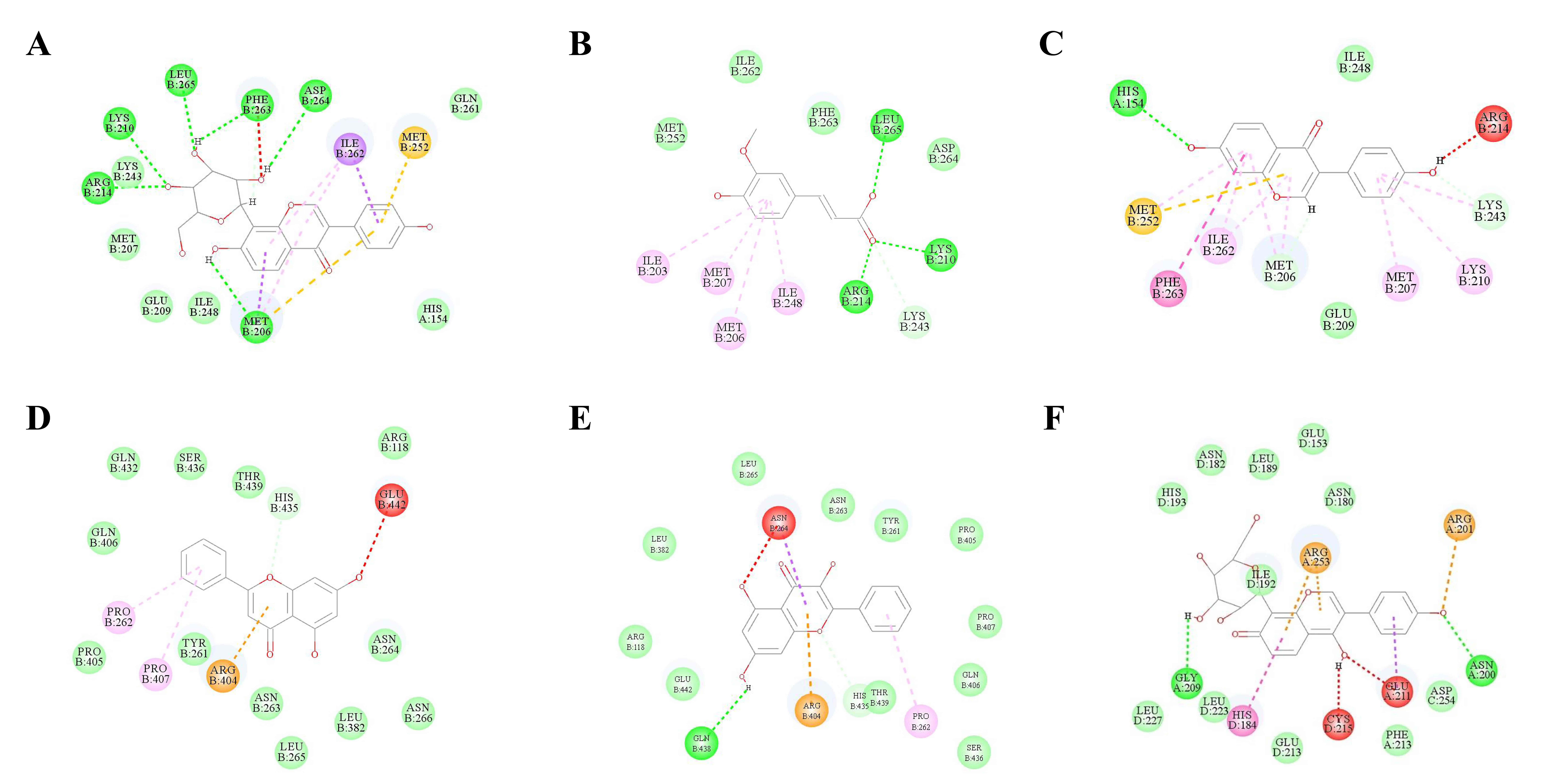 Fig. 5.
Fig. 5.Docking results of key target proteins with their corresponding compounds. (A) RELA and puerarin interaction diagram. (B) RELA and ferulic acid interaction diagram. (C) RELA and daidzein interaction diagram. (D) IKBKB and chrysin interaction diagram. (E) IKBKB and galangin interaction diagram. (F) NFKBIA and puerarin interaction diagram.
The GEO expression array data were used to investigate the proportion of
infiltrated immune cells in the peripheral blood of MI patients. All of the 41
samples met the conditions of CIBERSORT (p
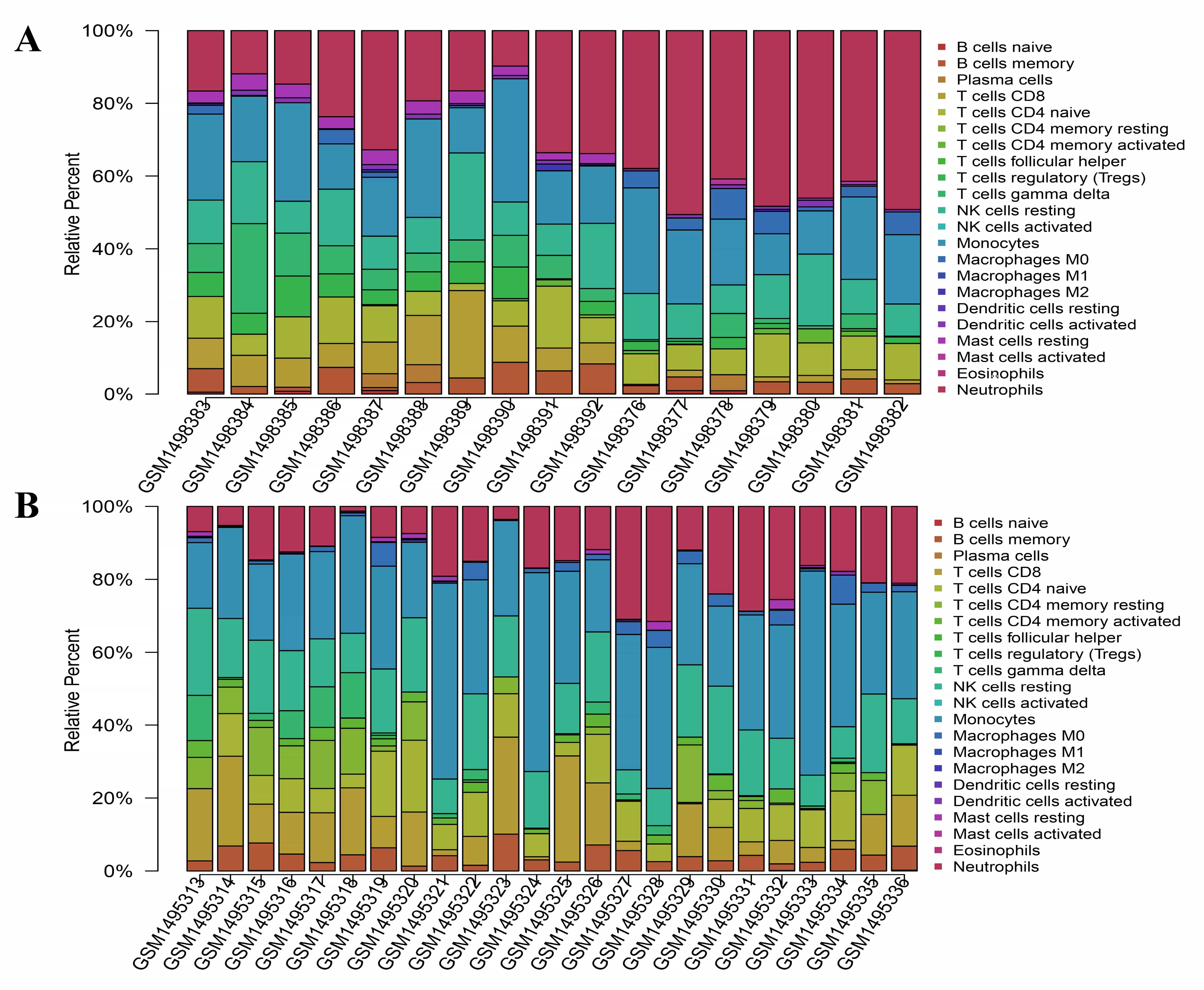 Fig. 6.
Fig. 6.The ratio of 22 immune cell subgroups in the peripheral blood of normal and MI patients. (A) GSE61145 expression array data set. (B) GSE60993 expression array data set. X axis: each GEO sample; Y axis: the percentage of each type of immune cell.
By utilizing network pharmacology screening and a literature review to obtain
the high-content medicinal ingredients and then according to the “Chinese
Pharmacopoeia” 2020 edition regulations, puerarin, daidzein, 3’-methoxydaidzein
in P. lobate, dioscin in D. nipponica Makino,
tetramethylpyrazine and ferulic acid in L. wallichii, and chrysin and
galangin in propolis were selected as quality control standard samples. First, 1
mL of concentrated CLMN solution was volatized to 5 mL in methanol to obtain the
test solution. puerarin, daidzein, 3’-methoxydaidzein, dioscin,
tetramethylpyrazine, and ferulic acid, chrysin, and galangin reference substances
were accurately weighed and placed in a 25-mL volumetric flask, dissolved in a
solution of phosphoric acid-water:acetonitrile = 95:5, shaken well, and the
volume was fixed to obtain the mixed reference substance solution. All of the
samples were analyzed on an Agilent column (4.6
| Time | Phosphoric acid - water/% | Acetonitrile/% |
| 0 | 95 | 5 |
| 9 | 93 | 7 |
| 27 | 90 | 10 |
| 45 | 87 | 13 |
| 50 | 85 | 15 |
| 65 | 76 | 24 |
| 73 | 65 | 35 |
| 85 | 49 | 51 |
| 98 | 49 | 51 |
| 110 | 35 | 65 |
| 115 | 95 | 5 |
| 130 | 95 | 5 |
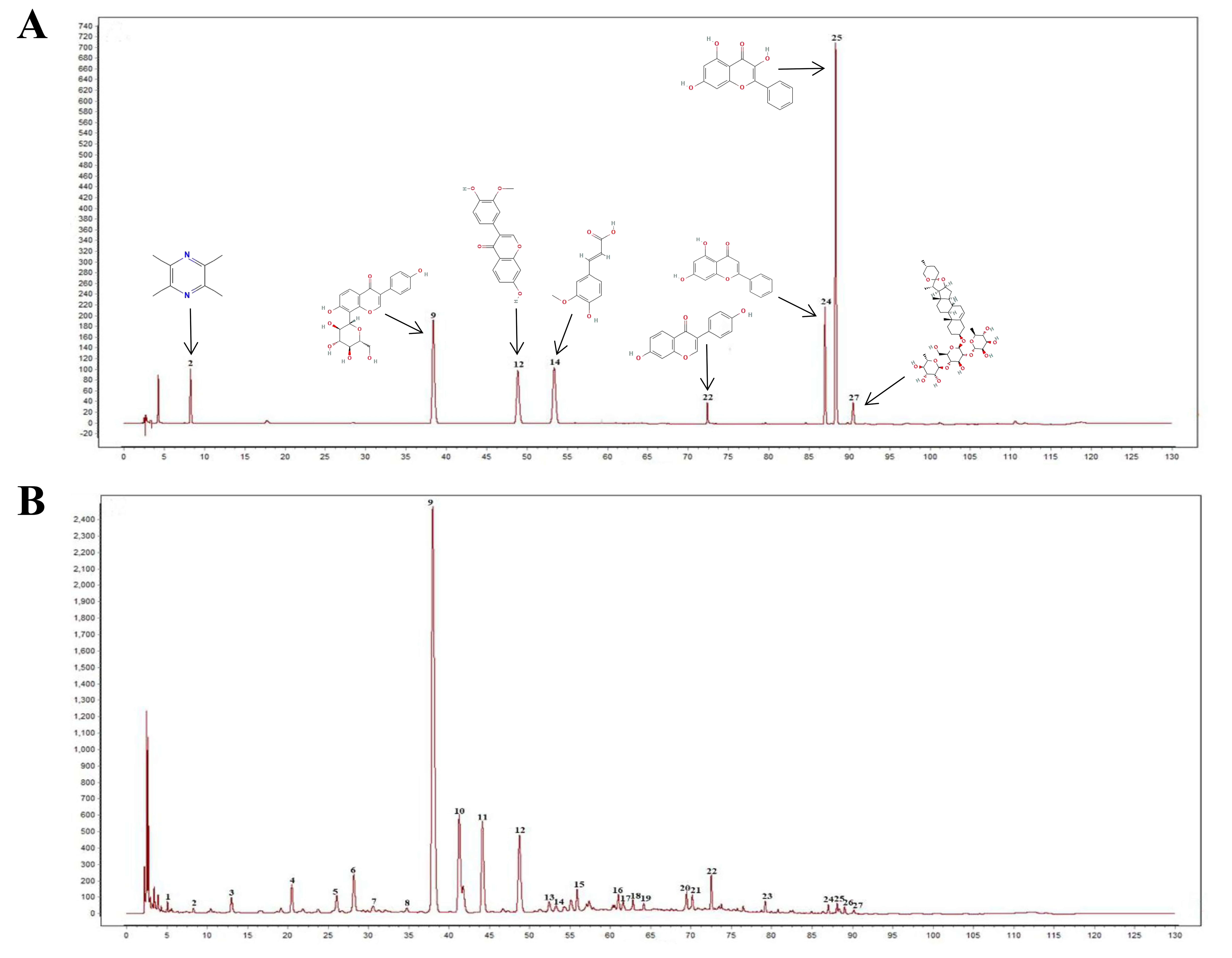 Fig. 7.
Fig. 7.Quality control of CLMN. (A) mixed reference substance. (B) CLMN fingerprint (No. 2: tetramethylpyrazine; No. 9: puerarin; No. 12: 3’-methoxydaidzein; No. 14: ferulic acid; No. 22: daidzein; No. 24: chrysin; 25: galangin; 27: dioscin).
A total of 8 mg of puerarin, 1.6 mg of ferulic acid, 1.6 mg of tetramethylpyrazine, 0.48 mg of 3’-methoxydaidzein, 0.08 mg of chrysin, 0.16 mg of galangin, 0.16 mg of dioscin, and 1.6 mg of daidzein were accurately weighed and placed in a 10 mL volumetric flask. Phosphoric acid-water: acetonitrile = 95:5 solution standard, was added to a constant volume, shaken well, and set aside. The mother liquor 1 was absorbed into 0.5, 0.25, 0.125, 0.0625, and 2 mL volumetric flasks, and the volume was adjusted using phosphoric acid-water, acetonitrile = 95:5 solution to the scale, and then shaken well. Low to high concentrations of the mixed reference solution were injected into the liquid chromatograph, and the peak areas were recorded. The x-coordinate (X) was the concentration, and the y-coordinate (Y) was the peak area. The regression equation of each reference is shown in Table 3 below.
| Reference substance | Regression equation | R |
Linear range ( |
| Puerarin | Y = 0.2017 X-0.8851 | R |
25 |
| Tetramethylpyrazine | Y = 1.4085 X-0.3883 | R |
5 |
| Ferulic acid | Y = 0.6873 X-0.1697 | R |
5 |
| 3’-methoxydaidzein | Y = 6.5348 X-4.5597 | R |
3 |
| Daidzein | Y = 2.0826 X-0.4641 | R |
5 |
| Chrysin | Y = 8.1272 X-0.3142 | R |
0.25 |
| Galangin | Y = 11.579 X-1.0619 | R |
0.5 |
| Dioscin | Y = 4.7049 X-1.6344 | R |
0.5 |
Samples to be tested were collected and determined according to the method under “4.9.1” to obtain the content of the effective components of the CLMN. The results are shown in Table 4 below.
| Components | Content (mg/g) |
| Puerarin | 30.1145 |
| Tetramethylpyrazine | 0.1874 |
| Ferulic acid | 0.1796 |
| 3’-methoxydaidzein | 8.1933 |
| Daidzein | 1.2249 |
| Chrysin | 0.8966 |
| Galangin | 0.9688 |
| Dioscin | 0.3522 |
The results of the HE staining are shown in Fig. 8. The structure of the cardiomyocytes in the sham operation group was normal and orderly, with no pathological changes. However, in the MI model group, the cardiomyocytes were injured and disordered, with a large number of inflammatory cell infiltrates, and large areas of myocardial infarction and myocardial fibrosis. With the low dose of CLMN, more inflammatory cell infiltrates were observed. However, in the CDDP group, with the middle and high dose groups of CLMN, inflammation was alleviated, the area of myocardial infarction was significantly reduced, and cardiomyocyte fibrosis was inhibited.
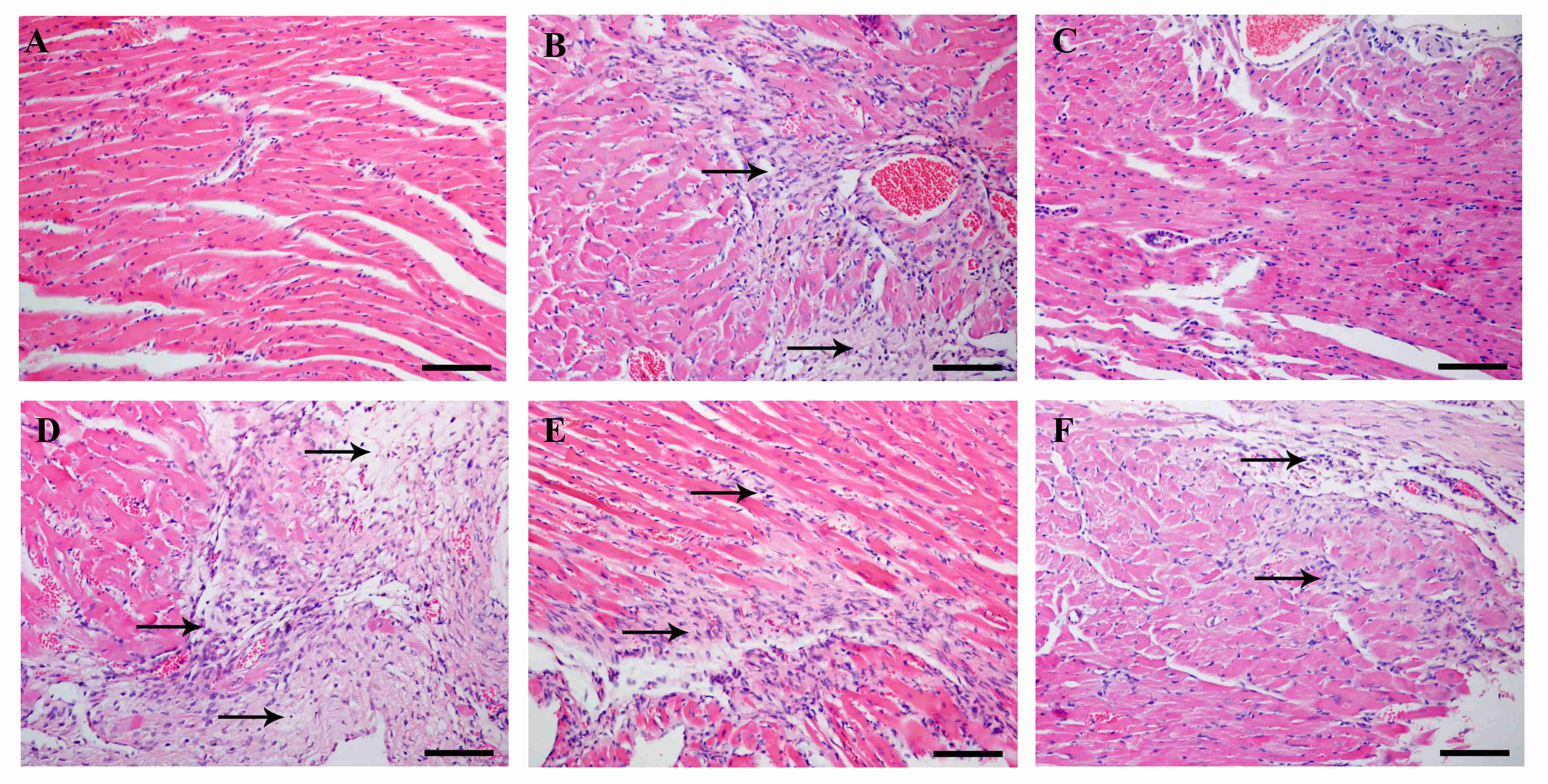 Fig. 8.
Fig. 8.The effect of CLMN on the pathomorphology of myocardial
tissue in rats (HE,
The results of the immunohistochemical staining (Fig. 9) showed that compared
with the sham group, the expression of the TNF-
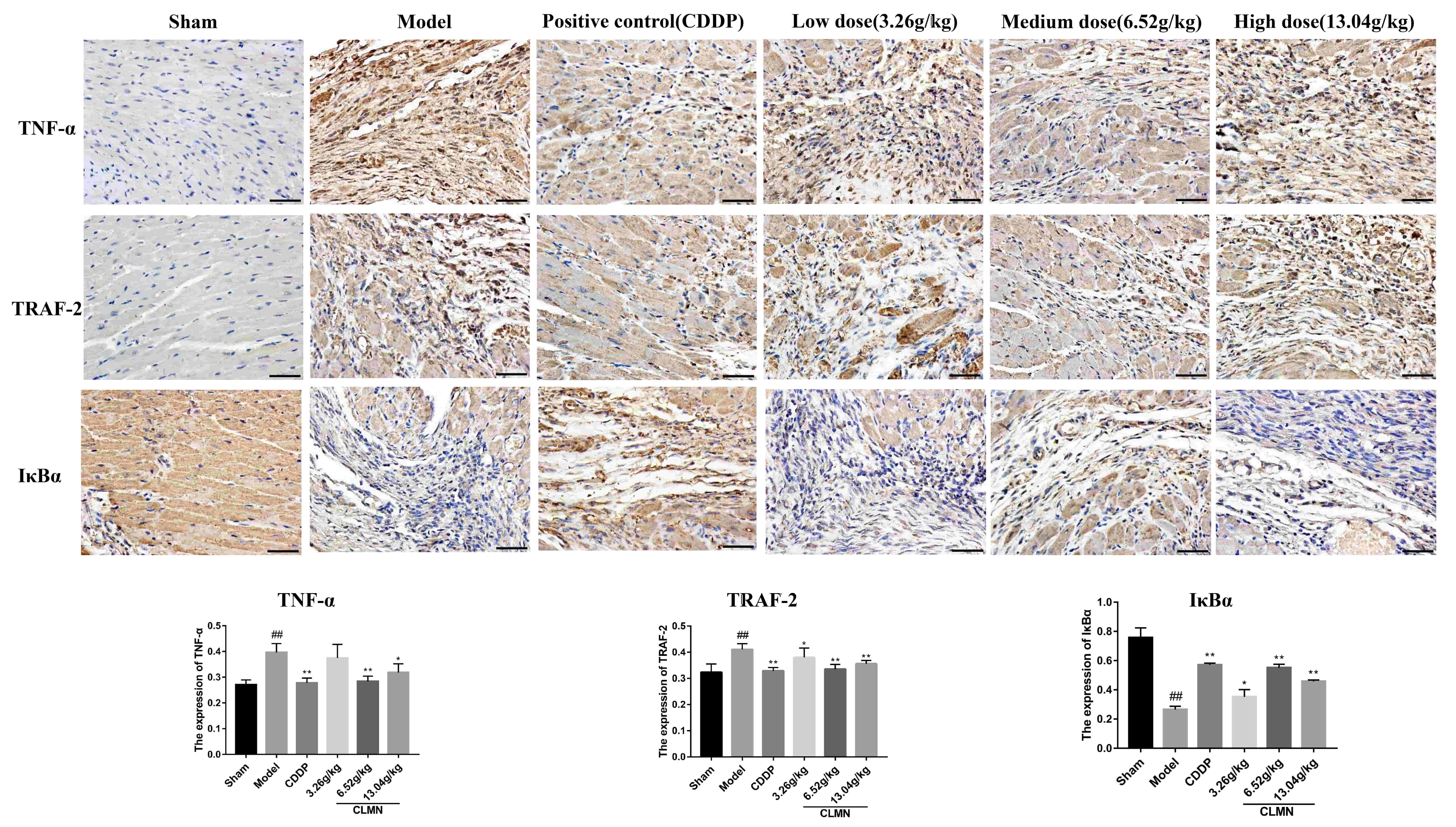 Fig. 9.
Fig. 9.
Effects of CLMN Pretreatment on the expression of
TNF-
MI causes an inflammatory response mediated by cytokines [28], that may decrease
myocardial contractile function and lead to myocardial hypertrophy, fibrosis and
remodeling. The study is essential to repair an infarcted heart. Therefore, the
regulatory mechanism of the inflammatory response urgently needs to be clarified,
and its mechanism of action is still under exploration. In this study, based upon
network pharmacology combined with the GEO chip, 52 core targets of CLMN were
screened, and the results of the docking between the core targets and active
components selected by the molecular docking software were stable. An MI mouse
model was established, and the three targets of TNF, TRAF-2,
and NFKBIA on the classical NF-Kappa B inflammatory signaling pathway
were verified using the immunohistochemistry method. We demonstrated that
TNF-
A PPI network analysis showed that MAPK1, STAT3,
MMP9, MAPK14, and other target genes had large degrees of
freedom, and these may be the key targets of CLMN in the treatment of MI. Target
proteins appear to be related and combine to produce the synergistic
effect of CLMN. MAPK1 is involved in a variety of cellular processes
such as proliferation, differentiation, and transcriptional regulation [29]. Zhao
et al. [30] showed that berberine down-regulates the p38MAPK-mediated
NF-Kappa signaling pathway, thereby increasing the expression of inflammatory
cytokines. Previous studies have indicated that phosphorylated STAT3
activates the NF-Kappa B signaling pathway, and the transcription factor, NF-KB,
enters the nucleus from the cytoplasm, thus regulating the expression of
inflammatory cytokines [31]. MMP9 is primarily regulated by the gene
transcription level. There are multiple transcription factor binding sites, such
as nuclear factor NF-KB, in the promoter of the MMP9 gene [32], and
NF-KB can enhance the expression of MMP9, which in turn plays an
important role in the occurrence and development of coronary heart disease [33].
A large number of studies have shown that MAPK14 activity plays a
crucial role in the body’s inflammatory response and is closely related
to cardiovascular disease. When MAPK14 is activated, it initiates an
inflammatory cascade, further phosphorylates the downstream protein kinases and
transcription factors, and then up-regulates the expression of
TNF-
The biological process enrichment analysis of CLMN showed that
target-related biological processes areprimarily involved in the
cellular response to chemical stress, the response to oxidative stress, and the
regulation of reactive oxygen species metabolic process. The enrichment pathway
of KEGG suggested the important involvement of Th17 cell differentiation, NF-
Kappa B and other inflammatory signaling pathways, which supports the observation
that the pathogenesis of MI is related to inflammation. TNF-
The CIBERSORT algorithm and ggplot2 package analysis showed that neutrophils,
macrophages, T cells, and other immune cells played a key role in the development
of MI. Neutrophils play a dual role in the pathophysic process following MI. In
the early stage after MI, the pro-inflammatory type neutrophils have a strong
pro-inflammatory and pro-injury effect. As time goes by, the proportion of the
anti-inflammatory type increases, participating in the “damage repair response”
after MI and playing an anti-inflammatory and anti-injury role [41]. Macrophages
are one of the major cell types involved in the inflammatory response after
myocardial infarction, and they can regulate heart regeneration by triggering the
inflammatory response. Studies have shown that after heart damage, neonatal mice
can selectively expand MHC-II
A certain chemical component of Chinese medicine cannot reflect the quality of Chinese medicine. All Chinese medicine acts through a synergistic effect of multiple components and targets. In this study, the quality of Chinese medicine was evaluated using the method of content determination. In this study, we screened eight potential pharmacodynamic components in the CLMN decoction, including puerarin, daidzein, 3’-methoxydaidzein, dioscin, tetramethylpyrazine, ferulic acid, chrysin, and galangin. The quality standard of CLMN was established, and the eight effective components were determined. The content determination of the multi-index components in the CLMN decoction established in this experiment can be used for quality control of the substance reference of CLMN.
In summary, this study systematically studied the mechanism of CLMN for the treatment of MI. CLMN may regulate the NF-Kappa B signaling pathway and the expression of RELA, IKBKB, NFKBIA, and other targets in the pathway for treating MI through the active components such as puerarin, daidzein, ferulic acid, galangin, and chrysin. CLMN treatment of MI is a process involving multiple-components, multiple targets, and multiple pathways. This study not only provides a new explanation for the “multi-component, multi-target, and multi-pathway” effect of CLMN treatment of MI, but also screens out some major active compounds of CLMN and establishes the quality standard of the CLMN decoction. In addition, the CIBERSORT algorithm was also used to evaluate the immune cell infiltration in MI, and the three targets of TNF-a, TRAF-2, and NFKBIA on the classic inflammatory pathway NF-Kappa B pathway were verified. The study demonstrated that TCM, CLMN in particular, has diverse pharmacologically active components, that may act synergistically to provide treatment for MI.
In this study, network pharmacology and experimental validation were combined to
explore the potential mechanism of CLMN in the treatment of MI. CLMN can
effectively decrease the expression of TNF-
JL, XW and DB performed the data analysis, wrote the first version of the manuscript and processed the graph and the table in the manuscript. JBZ, SX, YW and YJ finalized the manuscript. SY, WW, JHZ and JH collected the data. XZ and CW (corresponding author) conceived and coordinated the study. All authors read and approved the final manuscript.
The experimental animals were obtained with the informed consent of all participants. The institutional review board of the Shaanxi University of Chinese Medicine approved this experimental, code SYXK (chuan) 2020-030.
Thanks to all the peer reviewers for their opinions and suggestions.
This work was supported by Engineering and Technology Research Center for Application and Development of Chinese Herbal Medicine in Qinling, Shaanxi Province (20082DGC-32), the National Natural Science Foundation of China (Grant no. 81373944), Discipline Innovation team Project of Shaanxi University of Chinese Medicine (2019-YL11), Shaanxi University of Traditional Chinese Medicine, the key technology innovation team for the integration of traditional Chinese medicine (2018TD-005), Scientific Research in the affiliated Hospital of Shaanxi University of Chinese Medicine (2020ZJ005) and Shaanxi Provincial Key Research and Development Program (2017SF-351).
The authors declare no conflict of interest.
MI, Myocardial Infarction; CLMN, Compound Longmaining; HE, hematoxylin-eosin; TCMs, Traditional Chinese medicines; DEGs, differentially expressed genes; TCMSP, Traditional Chinese Medicine Systems Pharmacology; OB, Oral Bioavailability; DL, Drug-likeness; PPI, protein-protein interaction; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
