† These authors contributed equally.
Background: The coronavirus disease 2019 pandemic, caused by the severe
acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected more than 210
million individuals globally and resulted in over 4 million deaths since the
first report in December 2019. The early use of traditional Chinese medicine
(TCM) for light and ordinary patients, can rapidly improve symptoms, shorten
hospitalization days and reduce severe cases transformed from light and normal.
Many TCM formulas and products have a wide application in treating infectious and
non-infectious diseases. Polygonum cuspidatum Sieb. et Zucc. (P.
cuspidatum), is an important Traditional Chinese Medicine with actions of
clearing away heat and eliminating dampness, draining the gallbladder to relieve
jaundice, removing blood stasis to alleviate pain, resolving phlegm and arrest
cough. In the search for anti-SARS-CoV-2, P. cuspidatum was recommended
as as a therapeutic drug of COVID-19 pneumonia.In this study, we aimed to
identifies P. cuspidatum is the potential broad-spectrum inhibitor for
the treatment of coronaviruses infections. Methods: In the present study
, we infected human malignant embryonal rhabdomyoma (RD) cells with the OC43
strain of the coronavirus, which represent an alternative model for SARS-CoV-2
and then employed the cell viability assay kit for the antiviral activity. We
combined computer aided virtual screening to predicte the binding site and
employed Surface plasmon resonance analysis (SPR) to comfirm the interaction
between drugs and coronavirus.
We employed fluorescence resonance energy transfer technology to identify drug’s
inhibition in the proteolytic activity of 3CLpro and Plpro. Results:
Based on our results, polydatin and resveratrol derived from P.
cuspidatum significantly suppressed HCoV-OC43 replication. 50% inhibitory
concentration (IC
An unusual pneumonia of unknown origin was reported in December 2019 [1]. Its clinical features are similar to those of severe acute respiratory virus (SARS) [2]. The viral genome isolated from patients clustered into a clade of betacoronaviruses was distinct from that of the severe acute respiratory syndrome coronavirus (SARS-CoV) [3]. Thus, this disease, named the coronavirus disease 2019 (COVID-19) was caused by a novel coronavirus (2019-nCoV), which was later renamed the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). On March 11, 2020, COVID-19 was declared a global pandemic by the World Health Organization (WHO) [4]. In December 2020, several vaccines shown to be highly effective were granted emergency use authorization. While vaccines will prevent disease occurrence, infected individuals still need treatment options, and repurposing drugs circumvents the lengthy and costly process of drug development. Therefore, it is urgent to excavate the efficacy of currently approved drugs for the treatment of COVID-19 [5, 6, 7].
Traditional Chinese medicine (TCM) is an important party of the world complementary and alternative medicine. TCM offers a wide variety of application of traditional medicines and herbs for treating human diseases. Treating epidemic diseases with TCM and the treatment scheme of integrated Chinese and western medicine have proven their effectiveness in clinical practice in the management of COVID-19 pneumonia [8, 9, 10]. Effective prescriptions for the treatment of COVID-19 pneumonia include Qingfei Paidu decoction, Pneumonia No.1 Formula, and Shufeng Jiedu capsule. Polygonum cuspidatum Sieb. et Zucc., commonly known as Huzhang in Chinese and Japanese knotweed in English, is a widely used medicinal plant. The roots of Polygonum cuspidatum are used as an important traditional Chinese medicine for centuries, which have the actions of clearing away heat and eliminating dampness, draining the gallbladder to relieve jaundice, removing blood stasis to alleviate pain, resolving phlegm and arrest cough [11]. Of the traditional medicines tested, Polygonum cuspidatum and its active components, resveratrol, were found to inhibit Middle East respiratory syndrome coronavirus (MERS-CoV) in Vero E6 cells [12]. Over 67 compounds have been identified in P. cuspidatum, including quinones, stilbenes, flavonoids, coumarins, and lignans [13]. P. cuspidatum and its active components, resveratrol and emodin, were also found to attenuate influenza viral replication in A549 cells [14]. During the fight against COVID-19 in China, Chinese scientists found some herbs to be symptomatic for COVID-19 through clinical practice and through component screening in the Chinese medicine component library. One of them is P. cuspidatum, in which Polydatin has the strongest inhibitory effect on coronaviruses [15]. Hence, P. cuspidatum was recommended by the academician Zhang boli in the treatment of COVID-19 pneumonia [16].
The first genome sequence of SARS-CoV-2 has been entered into the database by Wu et al. [17] in February 2020. Coronavirus has six open reading frames (ORFs) and the first ORF (ORF1a/b), which comprises approximately 67% of the entire genome, encodes 16 non-structural proteins [18]. The remaining ORFs encode four major structural proteins, which are spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), nucleocapsid protein (N), and accessory proteins.
The polypeptides are released from each polyprotein through extensive proteolytic processing, primarily performed by the virally encoded chymotrypsin-like protease 3CL protein (3CLpro, also called the main protease, Mpro) with additional cleavage performed by the viral papain-like protease Pl protein (Plpro) [19, 20]. Owing to their essential roles in viral replication, both the proteases are recognized as attractive targets for the development of anti-SARS-CoV-2 therapeutics [21, 22]. While searching for a SARS-CoV-2 3CLpro and PLpro inhibitor from natural sources, we found that polydatin and resveratrol from P. cuspidatum possessed inhibitory activity against both the proteases. Herein, we identified the active ingredients from P. cuspidatum and their mechanism of inhibiting coronavirus.
Polydatin and resveratrol were purchased from solarbio company
(Solarbio Science & Technology Company, Beijing, China). The
Trans1-T1 strain,
F-
HCoV-OC43(OC43) strain was purchased from American Type Culture Collection (Human Coronavirus OC43(ATCC VR-1558) and propagated using RD cells (ATCC CCL-136). OC43 stock virus was obtained in RD cells in DMEM supplemented with 2% FBS at 72 h post-infection (hpi). Viral titer (PFU/mL) was determined by plaque assay.
RD cells were infected OC43 at the multiplicity of infection
(MOI = 0.01 IU) in the presence of indicated compound diluted in DMEM
supplemented with 2% FBS [23]. The compounds and the virus were maintained with
the cells during the 48 h incubation at 37
The median cytotoxic concentration (CC
The SARS-CoV-2 3CLP (pdb:6lu7) and PLP (pdb:6w9c) program database (PDB) files were downloaded from the PDB website (http://www.rcsb.org/). All heterogeneous atoms and the 3CLP ligand were removed and 6lu7 chain A was selected for subsequent molecular docking. The 3CLP docking grid was maximized for polydatin docking. All heterogeneous atoms and the PLP ligand were removed and 6w9c chain A was selected for subsequent molecular docking. The PLP docking grid was maximized for polydatin docking.
PDB file (6lu7 chain A) and (6w9c chain A) were converted to the PDBQT format as
macromolecules before virtual screening. The grid (ligand docking search space)
was located as described above. Then, Autodock Vina 1.1.2 [26] was used for the
subsequent molecular docking. Protein–ligand interactions were visualized using
Pymol version 1.7.4.5 (Schrödinger, New York, NY, USA). The amino acid
residues of 3CLP protein close to the hit ligands (
To conform the binding affinities of polydatin and resveratrol to SARS-CoV-2,
SARS-CoV, MERS-CoV 3CLpro and PLpro, SPR
technology based Biacore T200 biosensor was used (Biacore AB, Uppsala, Sweden)
[27]. At 25
All the screening assays were performed over the unmodified dextran surface and
the protein surface. During the 3CLpro and PLpro binder assay, the sample
concentrations were set at 100
To further investigate possible inhibitory activities of polydatin and
resveratrol against SARS-CoV-2, SARS-CoV, and MERS-CoV 3CLpro, and
PLpro, the inhibitory effects of the compounds were tested by the FRET method
[28]. For this assay, the internally quenched fluorogenic substrate
Dabcyl-KNSTLQSGLRKEE-Edans and Dabcyl-KRLKGGAPIKGE-Edans substrate (synthesized
by genescript company) was applied and the compounds were diluted to 100 mM, 20
mM, 10 mM, 2 mM, 1 mM, 200
In the above formula, A
To verify the inhibitory effect of polydatin
and resveratrol on coronavirus at the cellular level in vitro, we
measured the antiviral activity of polydatin and resveratrol by indirect
immunofluorescence assay (IFA) on RD cells
infected with OC43-CoV which is an alternative model for SARS-CoV-2. As shown in
Fig. 1A and 1B, polydatin and resveratrol significantly suppressed HCoV-OC43
replication, compared with that of the control (DMSO), and with over 90%
inhibitory effect at a concentration of 100
 Fig. 1.
Fig. 1.Confirmation of anti-coronavirus activity by immunofluorescence assay (IFA) analysis. The IFA of the human coronavirus strain OC43 (HCoV-OC43) nucleocapsid (N) protein in inhibitor-treated RD cells. RD cells in 96-well plates were infected with HCoV-OC43-wild type (multiplicity of infection = 0.01 IU) in serial dilutions of tested compounds, with dimethyl sulfoxide used as the negative controls. At 48 h post infection, the cells were analyzed by IFA for N protein expression. Nuclei (blue) were stained with Hoechst 33342.
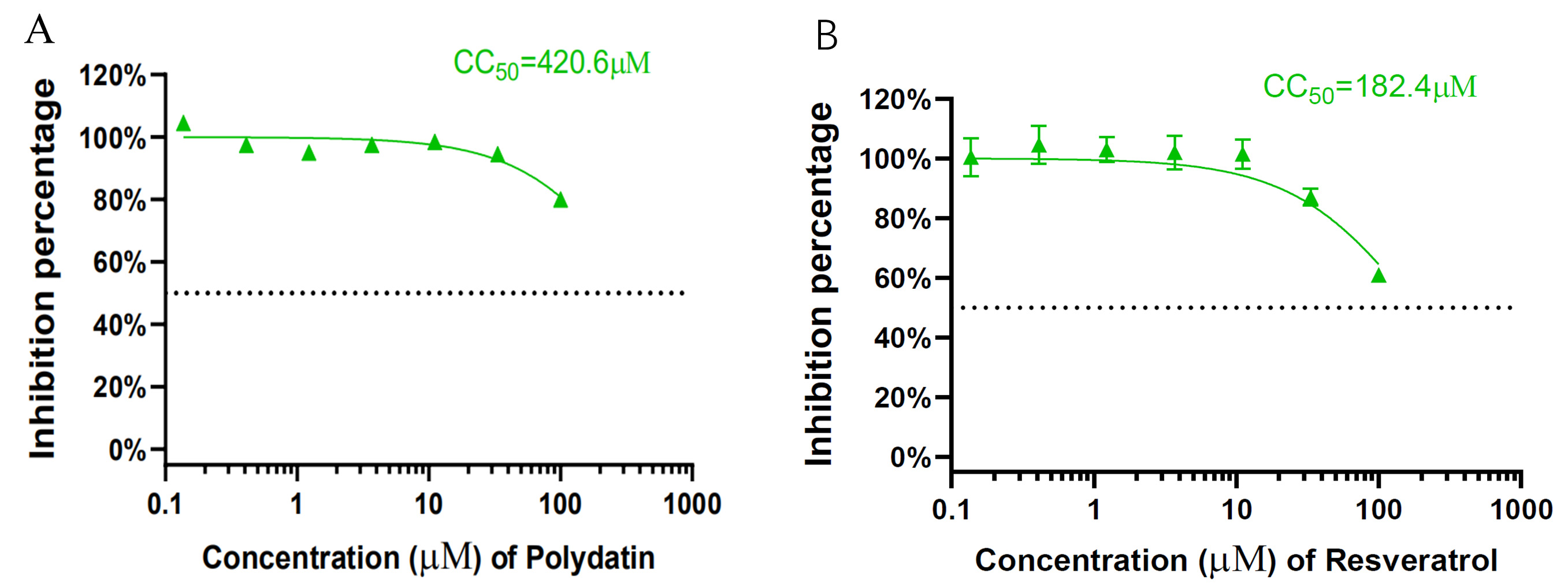 Fig. 2.
Fig. 2.Cytotoxicity of polydatin (A) and resveratrol (B) to HEK293T-ACE2 cells was measured by CellTiter-Blue assay. The median cytotoxic concentration value was determined by nonlinear regression analysis.
To elucidate the mechanism of polydatin and resveratrol on the inhibition of virus replication, we combined virtual screening with experimental studies. The chain A of 6lu7 (SARS-CoV-2 3CLP) was extracted to perform molecular docking with polydatin. For SARS-CoV-2 3CLP, polydatin binds to the PHE140, HIS163, MET165, GLU166, PRO168, and GLN189 amino acid sites (Fig. 3A). The chain A of 6w9c (SARS-CoV-2 PLP) was extracted to perform molecular docking with polydatin. For SARS-CoV-2 PLP, polydatin binds to the GLU67, GLU70, TYR71, PHE127, PRO130, and GLN133 amino acid sites (Fig. 3B).
 Fig. 3.
Fig. 3.Prediction of binding sites by virtual screening. (A) Polydatin binds to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) 3CLP (6lu7). Polydatin (gray stick) docked superposition on SARS-CoV-2 3CLP. Interactive amino acids (orange sticks) with polydatin (gray stick) on the SARS-CoV-2 3CLP (red dash line indicates the hydrogen bonds). (B) Polydatin binds to the SARS-CoV-2 PLP (6w9c). Polydatin (gray stick) docked superposition on the SARS-CoV-2 PLP, Interactive amino acids (orange sticks) with polydatin (gray stick) on the SARS-CoV-2 PLP (red dash line indicates the hydrogen bonds).
With the established FRET assay condition,
we tested polydatin and resveratrol to identify its inhibition in the proteolytic
activity of 3CLpro and PLpro (Table 1). Polydatin inhibited SARS-CoV-2 Mpro and
PLpro with 50% inhibitory concentration (IC
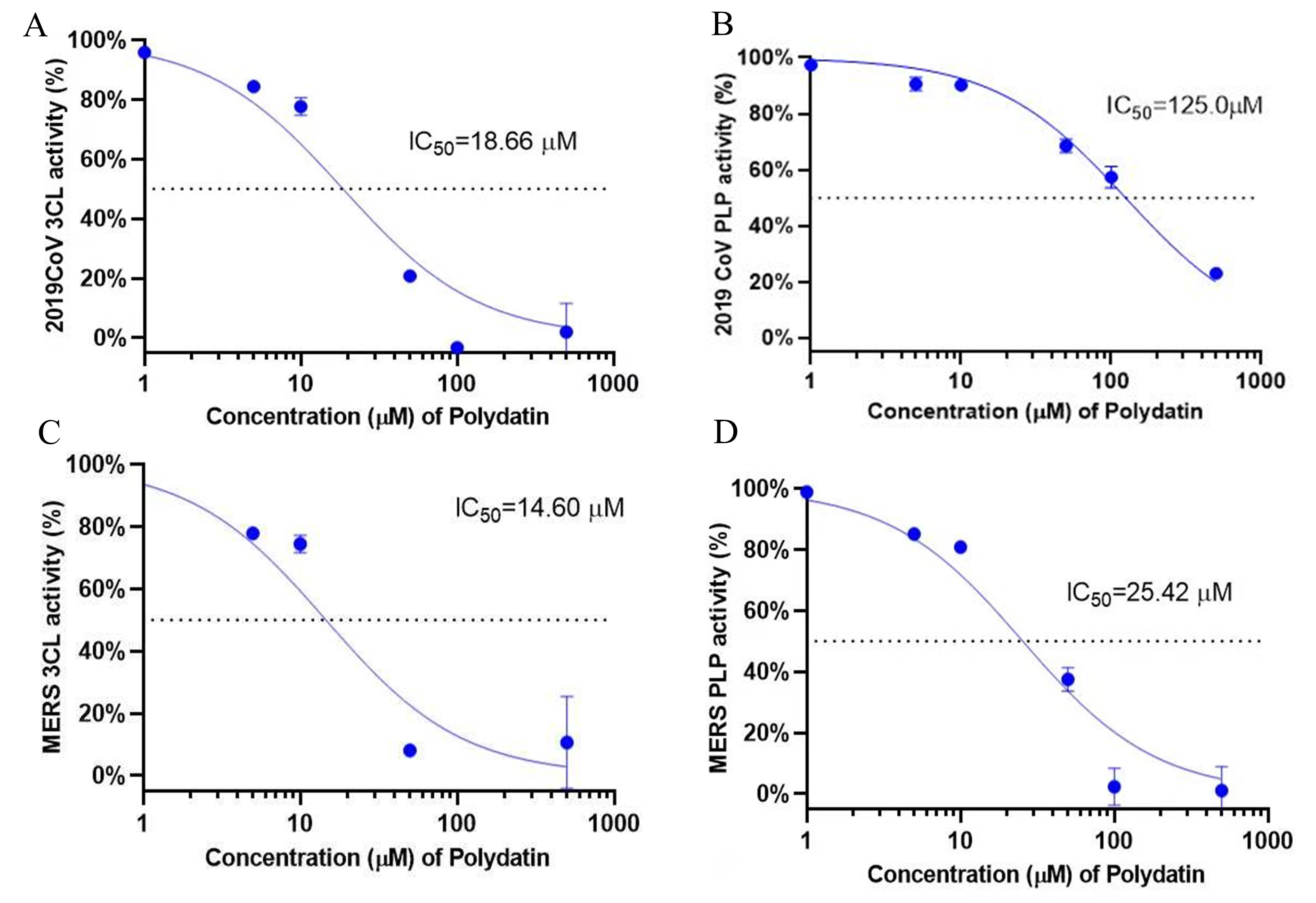 Fig. 4.
Fig. 4.
Determination of the 50% inhibitory concentration (IC
 Fig. 5.
Fig. 5.
Determination of the 50% inhibitory concentration (IC
| IC |
2019 CoV 3CL | 2019 CoV PLP | MERS 3CL | MERS PLP |
| Polydatin | 18.66 |
125 |
14.6 |
25.42 |
| Resveratrol | 29.81 |
60.86 |
16.35 |
19.04 |
| IC | ||||
To confirm the interaction between polydatin and resveratrol and SARS-CoV-2,
SARS-CoV, MERS-CoV 3CLpro, and PLpro protein, we tested whether
polydatin and resveratrol had high affinity to target protein using the SPR
assay. The SARS-CoV-2, SARS-CoV, and MERS-CoV 3CLpro and PLpro protein
were immobilized separately on a CM5 chip with the tested compounds flowing
across their surface. We found that
polydatin bound to SARS-CoV-2,
SARS-CoV, and MERS-CoV 3CLpro and PLpro protein exhibiting a strong
dose-dependent response, with respective KD (equilibrium dissociation constant)
values of 16.01
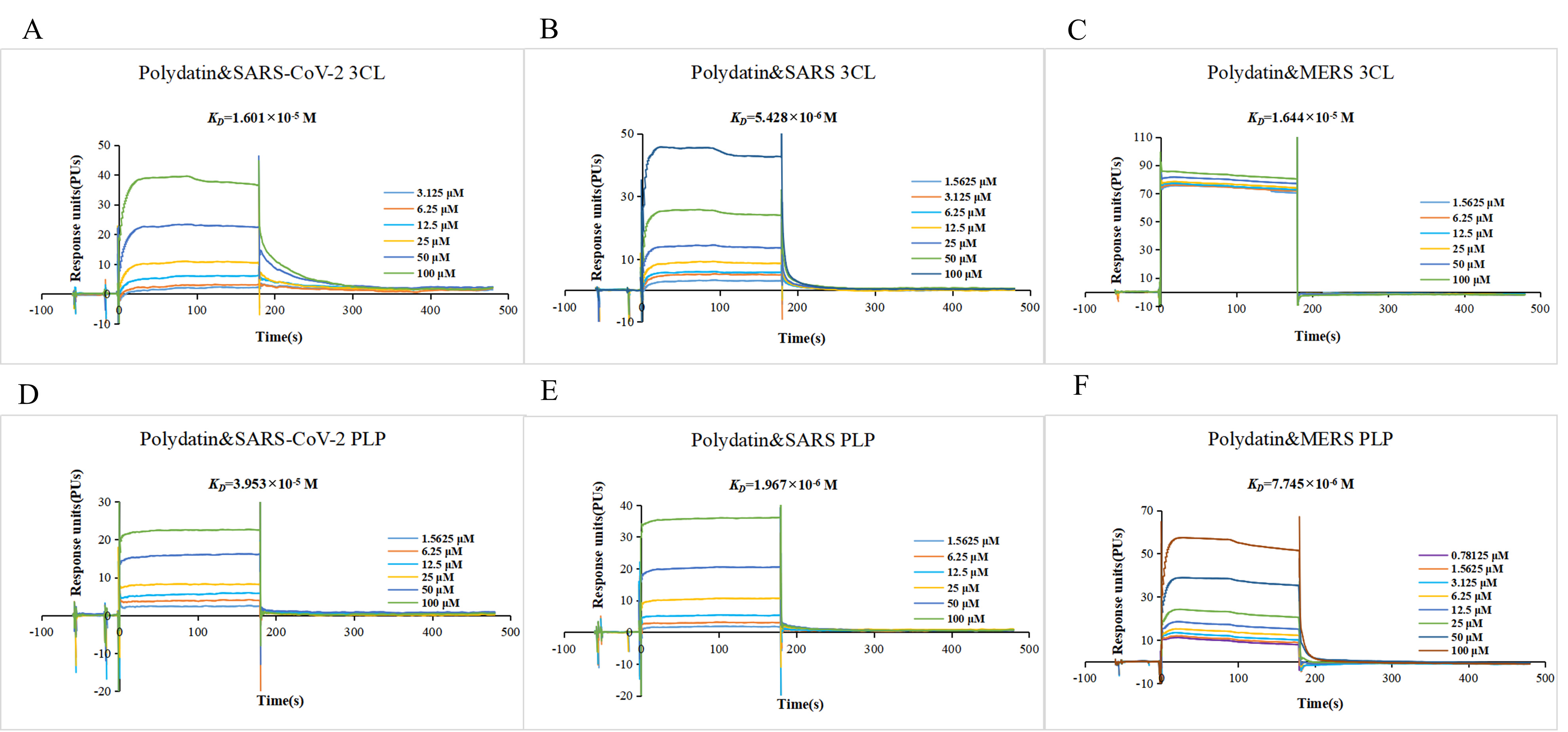 Fig. 6.
Fig. 6.Polydatin targets 3CLpro and PLpro of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) (A–F). The sensor image of 3CLpro and PLpro of SARS-CoV-2, SARS-CoV, and MERS-CoV with polydatin.
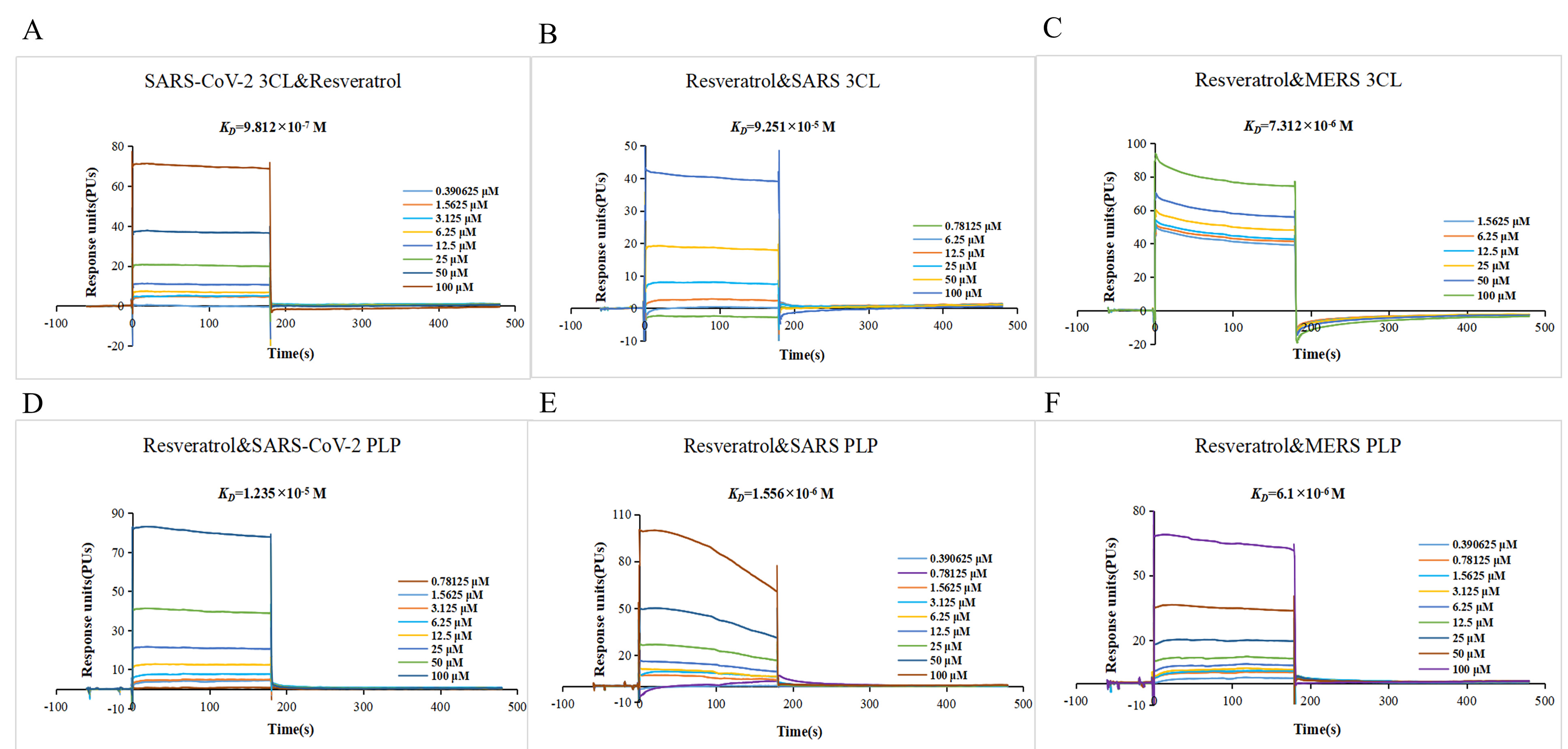 Fig. 7.
Fig. 7.Resveratrol targets 3CLpro and PLpro of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) (A–F). The sensor image of 3CLpro and PLpro of SARS-CoV-2, SARS-CoV, and MERS-CoV with resveratrol.
| Target | KD (M) | Rmax (RU) | Chi |
Chi |
| 2019 CoV 3CL | 1.601 × 10 |
14.1 | 1.35 | 1.16 |
| SARS 3CL | 5.428 × 10 |
9.251 | 0.313 | 0.56 |
| MERS 3CL | 1.644 × 10 |
6.966 | 0.0164 | 0.128 |
| 2019 CoV PLP | 3.953 × 10 |
16.65 | 0.00983 | 0.0992 |
| SARS PLP | 1.976 × 10 |
8.908 | 0.049 | 0.221 |
| MERS PLP | 7.745 × 10 |
7.163 | 0.386 | 0.621 |
| SARS-CoV-2, severe acute respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; PLP, papain-like protease; CL, chymotrypsin like protease. | ||||
| Target | KD (M) | Rmax (RU) | Chi |
Chi |
| 2019 CoV 3CL | 9.812 × 10 |
16.29 | 1.76 | 1.327 |
| SARS 3CL | 9.251 × 10 |
42.87 | 1.63 | 1.276 |
| MERS 3CL | 7.312 × 10 |
7.765 | 0.00146 | 0.0382 |
| 2019 CoV PLP | 1.235 × 10 |
26.02 | 0.217 | 0.466 |
| SARS PLP | 1.556 × 10 |
12.34 | 0.654 | 0.809 |
| MERS PLP | 6.100 × 10 |
13.78 | 0.739 | 0.86 |
| SARS-CoV-2, severe acute respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; PLP, papain-like protease; CL, chymotrypsin like protease. | ||||
Since the COVID-19 pandemic occurred in early 2020, scientists have been trying to find effective treatment methods and remedys. TCM has been proven effective for COVID-19 treatment [29, 30, 31]. P. cuspidatum, is a traditional Chinese medicine with actions of clearing away heat and eliminating dampness, draining the gallbladder to relieve jaundice, removing blood stasis to alleviate pain, resolving phlegm and arrest cough [32]. Polygonum cuspidatum and its active components were found to inhibit Middle East respiratory syndrome coronavirus (MERS-CoV) in Vero E6 cells [12] and attenuate influenza viral replication in A549 cells [14].
In this study, we focused on the SARS-CoV-2 3CL and PLP protein as target sites for the identification of possible Food and Drug Administration-approved drugs for repositioning in COVID-19 treatment.
As mentioned above, 3CLpro and PLpro of SARS-CoV-2 are good targets to design
inhibitory chemicals since some flavonoids inhibit the proteolytic activity of
SARS-CoV 3CLpro [33, 34]. The similar sequence between them [35, 36] together
with their conserved active site suggests that similar flavonoids may work for
SARS-CoV-2 3CLpro and PLpro. We found that polydatin and resveratrol revealed the
prominent inhibitory activity against SARS-CoV-2 and MERS-CoV 3CLpro and PLpro.
The measured IC
The COVID-19 pandemic wreaked havoc on human health from the beginning of 2020. In the search for anti-SARS-CoV-2 therapeutics, we identified the antiviral activity of flavonoids polydatin and resveratrol from P. cuspidatum on RD cells infected with the OC43 strain of the coronavirus. In order to elucidate the mechanism of polydatin and resveratrol on inhibition of virus replication, we combined virtual screening with experimental studies. Polydatin and resveratrol were found to be specific and selective inhibitors for SARS-CoV-2, 3CLpro and PLpro, viral cysteine proteases. In summary, this study identifies P. cuspidatum as the potential broad-spectrum inhibitor for the treatment of coronaviruses infections.
HX and NW wrote the manuscript, HX performed OC43 Viral infection inhibition
assay and determination of the median cytotoxic concentration and computer aided
virtual screening, JL and SS performed Surface plasmon resonance (SPR) analysis
and FRET technology–based IC
Not applicable.
Thanks to all the peer reviewers for their opinions and suggestions. We warmly appreciate professor Huan Yan (Wuhan University) for gifting OC43.
This work was partly supported by the National Science and Technology Major project (2017ZX09303008), Shenzhen Bay laboratory start up fund (21230071), Guangdong Provincial Special Projects on COVID-19 (2020KZDZX1182), Natural Science Foundation of China (61773196, 32070681), Guangdong Provincial Key Laboratory of Computational Science and Material Design (2019B030301001), Guangdong Provincial Key Laboratory of Cell Microenvironment and Disease Research (2017B030301018), Shenzhen Peacock Plan (KQTD2016053117035204), and Hebei Natural Science Foundation (H2017206281, H2020206483). Medical Science Research Project of Hebei Province (20211109). We thank MengSi.SUN (Office of Core Facilities, ShenZhen Bay Laboratory [SZBL]) for technical support for the SPR analysis. The Rapid Service Fees were funded by the authors.
The authors declare no conflict of interest.
