Objectives: To quantify the integrated levels of ACE2 and TMPRSS2, the two well-recognized severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry-related genes, and to further identify key factors contributing to SARS-CoV-2 susceptibility in head and neck squamous cell carcinoma (HNSC). Methods: We developed a metric of the potential for tissue infected with SARS-CoV-2 (“TPSI”) based on ACE2 and TMPRSS2 transcript levels and compared TPSI levels between tumor and matched normal tissues across 11 tumor types. For further analysis of HNSC, weighted gene co-expression network analysis (WGCNA), functional analysis, and single sample gene set enrichment analysis (ssGSEA) were conducted to investigate TPSI-relevant biological processes and their relationship with the immune landscape. TPSI-related factors were identified from clinical and mutational domains, followed by lasso regression to determine their relative effects on TPSI levels. Results: TPSI levels in tumors were generally lower than in the normal tissues. In HNSC, the genes highly associated with TPSI were enriched in viral entry-related processes, and TPSI levels were positively correlated with both eosinophils and T helper 17 (Th17) cell infiltration. Furthermore, the site of onset, human papillomaviruses (HPV) status, and nuclear receptor binding SET domain protein 1 (NSD1) mutations were identified as the most important factors shaping TPSI levels. Conclusions: This study identified the infection risk of SARS-CoV-2 between tumor and normal tissues, and provided evidence for the risk stratification of HNSC.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the pathogen causing Coronavirus disease 2019 (COVID-19) [1]. The virus has been disseminated worldwide and has severely threatened human health since its outbreak in Wuhan, China, in late 2019 [2]. Patients with COVID-19 often experience respiratory symptoms characterized by fever, cough, fatigue, and dyspnea, developing into multi-organ failure and even death in severe cases [3, 4]. Although SARS-CoV-2 infection primarily induces pulmonary involvement, extrapulmonary organ manifestations such as heart failure [5], renal dysfunction [6], and oral mucosal lesions [7] have also been observed. Multiple clinical studies also demonstrated that cancer patients have an increased risk of SARS-CoV-2 infection [8] and are prone to developing more complications [9].
Angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) are two well-recognized factors involved in SARS-CoV-2 cell entry [10, 11, 12]. To be specific, SARS-CoV-2 infection begins with the binding of the viral spike (S) protein to the ACE2 receptor, allowing viral adhesion to the surface of host cells [10, 11]. This binding is followed by TMPRSS2-enabling S-protein priming [12]. It was recently reported that additional host molecules including a disintegrin and metalloprotease 17 (ADAM17), cathepsin L (CTSL), and furin may also function as receptors for SARS-CoV-2. However, ADAM17 and CTSL have been shown to be nonessential for SARS-CoV-2 infection [13, 14]. Furin pre-activation can facilitate SARS-CoV-2 entry into some cells types, particularly those with low expression of TMPRSS2 or lysosomal cathepsins [15]. Several studies identified TMPRSS2 as an essential host cell factor for these respiratory viruses and further demonstrated that inhibition of virus activating host cell proteases [16, 17, 18] (e.g., TMPRSS2) provides a promising approach for developing therapeutics against treat respiratory virus infections. Camostat mesylate, a clinically proven serine protease inhibitor, can partially block SARS-2-S-driven entry into TMPRSS2+ Caco-2 and Vero-TMPRSS2 cells [19]. Nafamostat mesilate can inhibit Middle East respiratory syndrome (MERS)-CoV S protein-mediated viral membrane fusion with TMPRSS2- expressing Calu-3 lung host cells by inhibiting TMPRSS2 protease activity [20]. Because the S proteins of MERS-Cov and SARS-CoV-2 share considerable amino acid sequence homology [21], nafamostat mesilate may also inhibit SARS-CoV-2 cell entry.
Due to the important roles of ACE2 and TMPRSS2 in SARS-CoV-2 cell entry, researchers have investigated their expression to identify potential target organs or tissue types and predict the susceptibility to SARS-CoV-2 infection [22, 23]. However, current studies based on ACE2 and TMPRSS2 mainly analyze each factor separately. For instance, a recent study suggested that HNSCs are less likely to be infected with SARS-CoV-2 than normal tissues for the constant ACE2 but decreased TMPRSS2 [22]. Nevertheless, real individuals present unique combination of the two genes, and the heterogeneity of individual ACE2 and TMPRSS2 expression patterns appears to be ignored by this method. In particular, it is difficult to draw an exact conclusion when they change in opposite directions. To address these issues, a novel quantitative measure is needed to integrate both ACE2 and TMPRSS2 weights.
In 2015, Rooney et al. [24] proposed a method to quantitatively assess immune cytolytic activity using the geometric mean of the mRNA expression of two essential cytolytic effectors. Based on this approach, we constructed a metric of the potential for SARS-CoV-2 infection and calculated it for thousands of human cancer and matched normal tissue samples to further determine the susceptibility across diverse tissue types, focusing on viral entry-related genes. As the head and neck exert an important role in communicating with the outside world and the oral cavity is found to be a vital site for SARS-CoV-2 infection [25], we further performed a detailed analysis of head and neck squamous cell carcinoma (HNSC), establishing a correlation between the metric and various features including immune, clinical, and genomic domains.
The RNA sequencing (RNA-Seq) data included in the pan-cancer analysis were
acquired from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/).
Excluding tumors with less than 30 normal samples, the resulting analyzed cohorts
included 5624 primary tumors and 602 matched normal tissues (Fig. 1). The cancer
types consisted of breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD),
HNSC, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell
carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma
(LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD),
stomach adenocarcinoma (STAD), thyroid carcinoma (THCA). For each TCGA dataset, a
total of 19597 mRNAs were annotated according to Gencode V33. FPKM values were
converted to TPM and log
 Fig. 1.
Fig. 1.Distribution of both ACE2 and TMPRSS2 levels, and TPSI scores in
normal and tumor tissues across 11 tumor types in TCGA.
Further analysis of HNSC was conducted using the TCGA_HNSC dataset as a discovery set, which included 500 tumors and 44 normal tissues. Patients’ clinical information was retrieved from TCGA, and the human papillomaviruses (HPV) status was assessed at the Broad Institute based on DNA sequencing and PathSeq algorithm [26]. We also obtained the HNSC gene-level somatic mutations from UCSC Xena (https://xenabrowser.net/). To validate the main correlations of clinical and mutational features with our metric, the 500 HNSC patients were randomly divided into internal validation set-1 (n = 150) and set-2 (n = 350). Additional three datasets (GSE30784, GSE107591, and GSE41613) from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) were used as external validation sets. Among these sets, GSE30784 contained 167 HNSCs and 45 normal samples [27], GSE107591 contained 24 HNSCs and 23 normal samples [28], and GSE41613 had 97 tumor samples only [29]. The gene expression profiles were normalized through the normalizeBetweenArrays function in R package “limma”.
The potential for tissue infected with SARS-CoV-2 (TPSI) was computed as the
geometric mean of ACE2 and TMPRSS2 expression. In the TCGA_HNSC dataset, TPM
values were used to calculate TPSI, and transformed TPSI values log
In TCGA_HNSC, a total of 4899 mRNAs with the top 25% variance were selected to construct a co-expression network by the “WGCNA” package to identify modules and genes most strongly correlated with TPSI [30].
Gene ontology (GO) biological processes (BP) and KEGG enrichment analysis of the
related genes were conducted and visualized using ClueGO and CluePedia plugins
within Cytoscape software (version 3.7.2) [31]. A p-value
In this study, 13 pairs of surgically resected HNSCs and their adjacent normal
tissues were collected for qPCR from the School and Hospital of Stomatology,
China Medical University during July 2021. All samples were confirmed by
histopathology. The fresh tissues were immediately snap-frozen in liquid nitrogen
and stored at –80
Tissue RNA was isolated using TRIzol reagent (Takara) and reverse-transcribed
into cDNA using the PrimeScript
According to SARS-CoV-2 infection-related pathways (R-HSA-9694516) from Reactome (http://reactome.org/) and published literature [32, 33, 34], we generated three relevant host factor sets: (1) Other coronavirus entry-related factors (i.e., furin, ADAM17, CTSL, CTSB, BSG, TMPRSS4, TMPRSS11A, TMPRSS11B, ANPEP, CLEC4G, DPP4, and NRP1); (2) Translation of replicase and assembly of the replication transcription complex (i.e., CHMP2A, CHMP2B, CHMP3, CHMP4A, CHMP4B, CHMP4C, CHMP6, CHMP7, MAP1LC3B, BECN1, UVRAG, PIK3C3, and PIK3R4); (3) Replication of the SARS-CoV-2 genome (i.e., RB1, ZCRB1, VHL, DDX5, EEF1A1, MTHFD1). In addition, we accessed the marker genes for 28 subpopulations of tumor infiltrating immune cells [35]. TPM values of the genes in TCGA_HNSC were extracted for calculation. Corresponding SARS-CoV-2 infection-related and immune cell-type signature scores for individual HNSC samples were quantified using ssGSEA based on R package “GSVA” [36]. The immune cells were clustered by Euclidean distance and ward.D2 clustering method.
Differences in the distribution of ACE2 and TMPRSS2 expression, TPSI, and
signature scores between groups were evaluated using the Wilcox test. Chi-square
or Fisher’s exact test was used to compare categorical variables, as appropriate.
Spearman’s correlation analysis was performed to evaluate the correlation between
TPSI and other variables. The survival difference was assessed by Kaplan-Meier
analysis with the log-rank test. The independence test was conducted with the
coin package of R for identifying significant mutations related to TPSI levels.
The least absolute shrinkage and selection operator (LASSO) regression analysis
was used for qualitative assessment of variable importance by “glmnet” package
[37]. All statistical analyses were carried out in R software. The significance
level was set at two-tailed p
Considering the high prevalence of COVID-19 in cancer patients, we explored the distribution of ACE2 and TMPRSS2 expression in 11 cancer types and their corresponding normal tissues. Compared with normal tissues, the opposite trends in expression changes of ACE2 and TMPRSS2 were observed in KIRP and PRAD. Based on these observations, we could not determine the SARS-CoV-2 infection potential for the two tumor types relative to normal tissues. Subsequently, we devised a metric named TPSI to integrate the ACE2 and TMPRSS2 transcript levels in order to compare the susceptibility to SARS-CoV-2 infection between tumors and normal tissues. As shown in Fig. 1, the TPSI levels were highest in the colon and kidney and lowest in the breast. Moreover, 10 of 11 tumor types had lower TPSI levels than their matched normal tissues; only STAD presented the same levels for both groups.
To explore the genes highly correlated with TPSI in HNSC, we first constructed a WGCNA network from the TCGA_HNSC dataset. After outlier removal from 500 samples, a soft-threshold power of five was selected for identifying co-expression modules. As a result, the genes were classified into eight modules (Fig. 2A–C). The red module exhibited the strongest association with the TPSI score. It is worth mentioning that both ACE2 and TMPRSS2 were present in the red module. In particular, they were among the top 25% of the genes ranked according to the gene significance (GS) values used to determine how gene levels related to TPSI (Fig. 2C). We then carried out GO and KEGG enrichment analysis on 156 genes with GS greater than 0.2 in the red module. As anticipated, the viral infection-related process “regulation of viral entry into host cell” was significantly enriched (Fig. 2D).
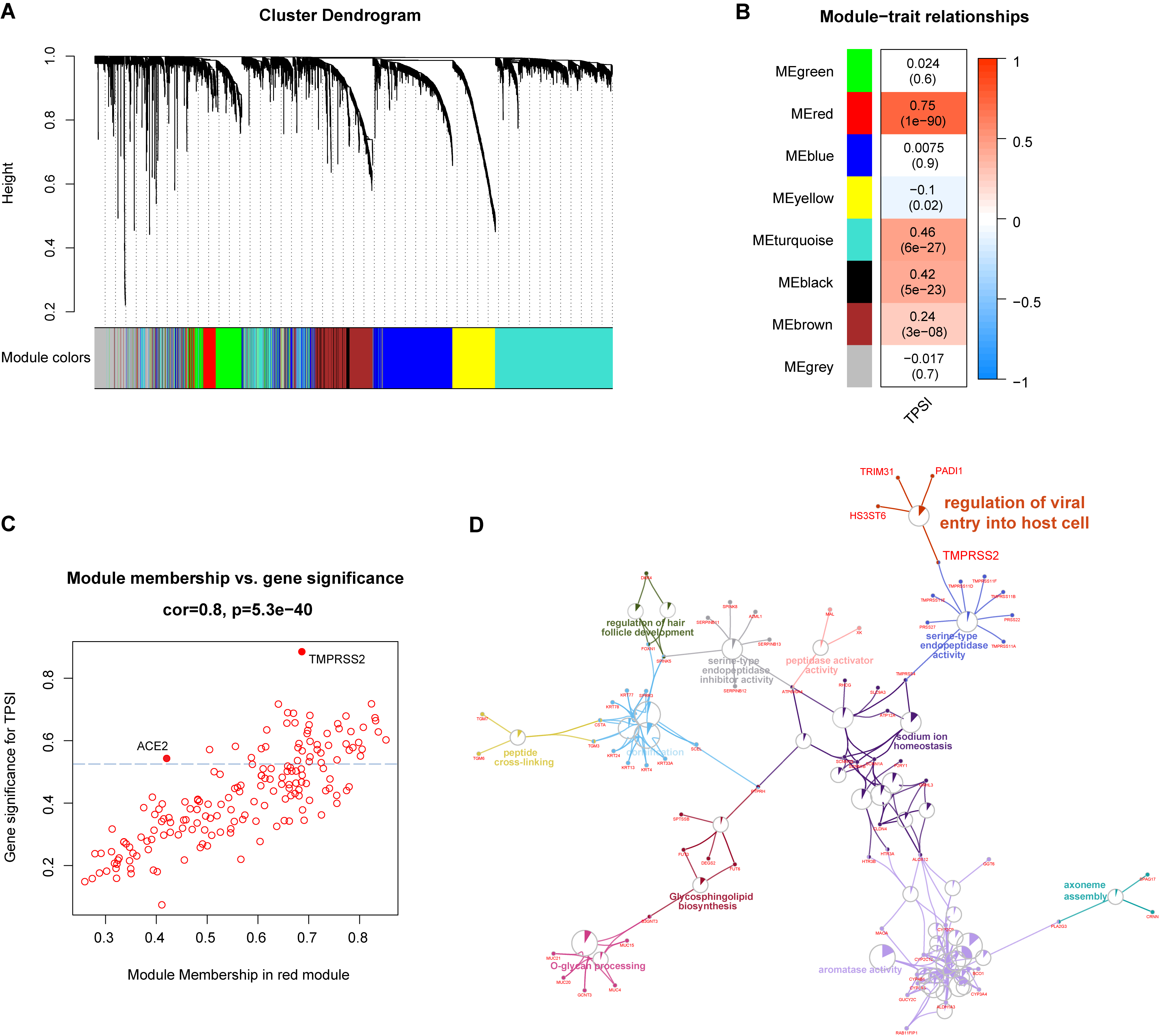 Fig. 2.
Fig. 2.WGCNA and functional analyses in the TCGA_HNSC dataset. (A)
The cluster dendrogram of genes with the top 25% variance. (B) Correlation
between module eigengenes and TPSI. (C) Scatter plot of genes in the red module;
scattered points above the horizontal blue dotted line represent the top 25%
genes with high GS. (D) GO and KEGG enrichment of the genes with GS
Two public datasets (GSE30784 and GSE107591) and our own clinical samples were used to verify the differences in the TPSI levels between HNSCs and normal tissues. All three datasets confirmed a lower TPSI level in the HNSCs (Fig. 3A,B). In addition, GSE41613 was used to validate the relationship between TPSI and viral entry-related processes. Significant positive correlations were observed between TPSI levels and both genes expression (Fig. 3C). The top 160 genes positively associated with TPSI were selected for functional enrichment, and the analysis revealed a significant enrichment of “entry into host cell” (Fig. 3D).
 Fig. 3.
Fig. 3.Validation of TPSI levels and TPSI-related functions. (A) Distribution of TPSI levels in normal tissues and HNSCs in GSE30784 and GSE107591. (B) Distribution of TPSI levels in normal tissues and HNSCs in our clinical sample sets. (C) Correlation between TPSI and both ACE2 and TMPRSS2 in GSE41613. (D) GO and KEGG enrichment of the top 160 genes positively associated with TPSI in GSE41613.
In addition to ACE2 and TMPRSS2, other coronavirus invasion-related molecules (e.g., furin) and virus replication may influence SARS-CoV-2 infection. Therefore, we investigated the relationship between relevant signatures and whether there are tumors or not. For the TCGA_HNSC dataset, ssGSEA was carried out to generate individual signature activity scores based on three SARS-CoV-2 infection-related gene sets. The results showed that “Other coronavirus entry-related factors” and “Translation of replicase and assembly of the replication transcription complex” were significantly downregulated in HNSCs compared to normal tissues, while there was no significant difference in “Replication of the SARS-CoV-2 genome” between the two groups (Fig. 4).
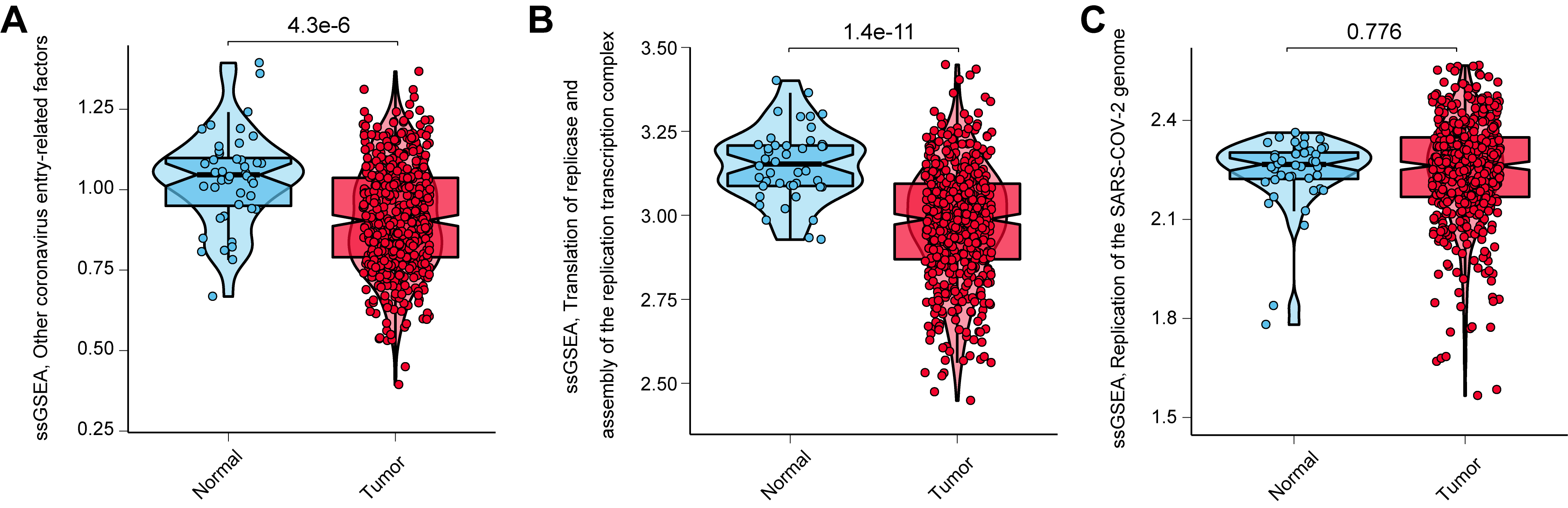 Fig. 4.
Fig. 4.Distribution of ssGSEA scores of other SARS-CoV-2 infection-related signatures in normal tissues and HNSCs in the TCGA_HNSC dataset. (A) Other coronavirus entry-related factors. (B) Translation of replicase and assembly of the replication transcription complex. (C) Replication of the SARS-CoV-2 genome.
Given the crucial roles that immune cells play in combating SARS-CoV-2 infection [38], we explored the potential associations between TPSI and tumor-infiltrating immune cells in HNSC. Based on the ssGSEA scores, 500 patients with HNSC were clustered into two immune infiltration subgroups: low (n = 175) and high (n = 325). The high immune infiltration group generally showed a higher abundance of 28 immune cell populations (Fig. 5A). The consistency of the abundance between immune cells exerting anti-tumor activity (e.g., activated CD8+T cells) and repressing such reactivity (e.g., regulatory T cells) might be partially ascribed to a feedback mechanism that anti-tumor inflammation promotes the recruitment or differentiation of immunosuppressive cells [39].
HPV is a pathogenic factor for HNSC. HPV+ and HPV- HNSCs are regarded as two radically different cancers, exhibiting distinct immune landscapes [40]. We found that the proportion of HPV+ or TPSI-high HNSC was higher in the high immune infiltration group. To further clarify the relationship between TPSI levels and immune infiltration, we compared the TPSI scores of the two immune infiltration subgroups in the HNSC cohort stratified by HPV status. Increased TPSI levels were observed in the high immune infiltration group for the HPV+ HNSCs; however, the difference was not significant for the HPV- HNSCs (Fig. 5B). These results indicated that there were no strong associations between overall immune infiltration and TPSI levels. We then measured TPSI correlations with different immune cells, and found that both T helper 17 (Th17) cells and eosinophils, known to mediate inflammation [41, 42], were positively correlated with TPSI levels in HNSC, without being affected by HPV stratification (Fig. 5C).
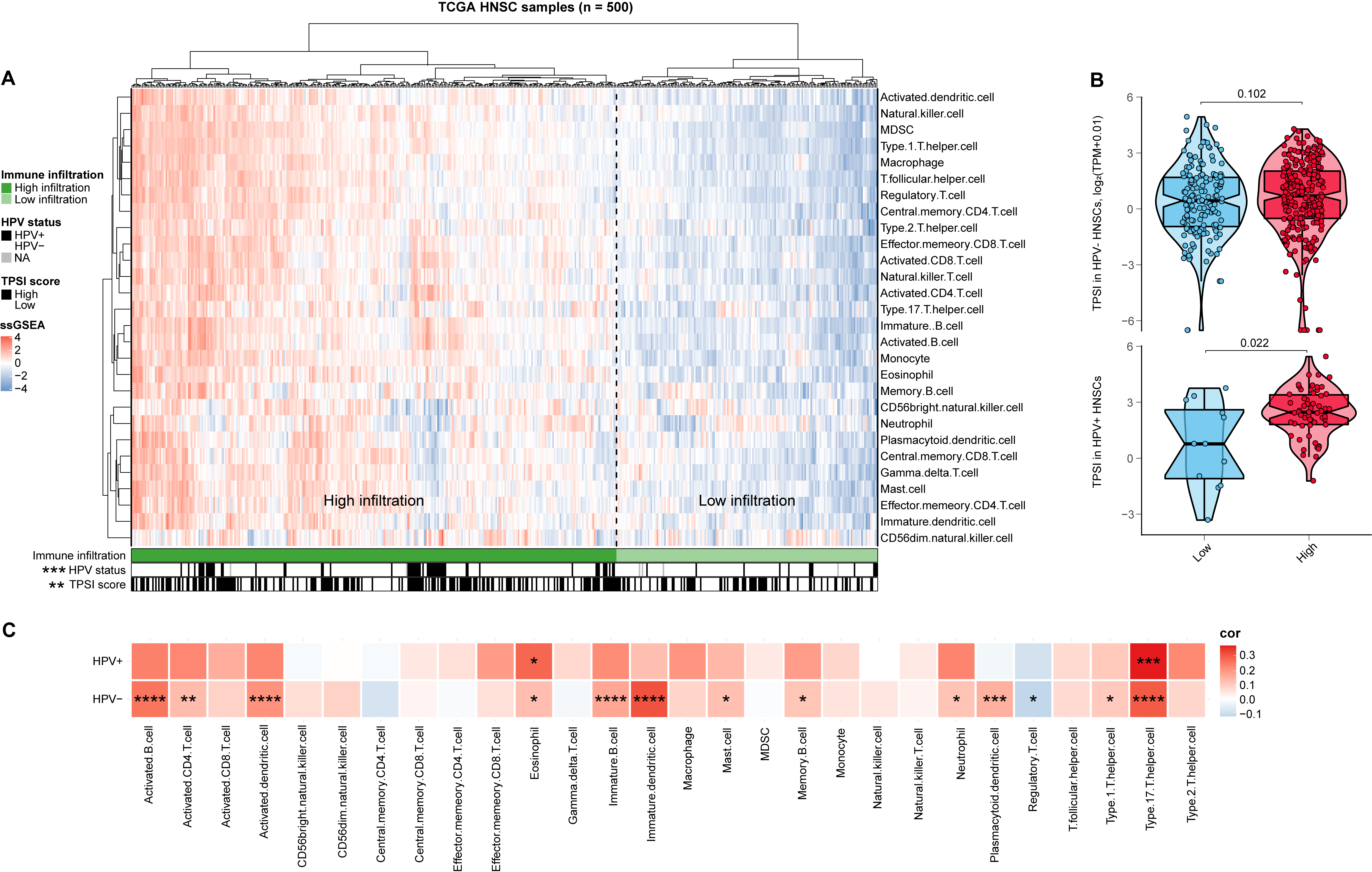 Fig. 5.
Fig. 5.Immune landscape of HNSC and its correlation with TPSI in TCGA_HNSC. (A) Heatmap of ssGSEA scores of 28 immune cell subpopulations. (B) Distribution of TPSI scores in low and high immune infiltration groups in HNSCs by HPV stratification. (C) Correlations between immune cells and TPSI in HNSCs by HPV stratification.
According to a number of clinical COVID-19 studies, clinical factors such as age
and gender might be related to infection and disease severity [43]. Thus, we
explored the correlations between various clinical features and TPSI levels in
TCGA_HNSC. The leading causes of HNSC include long-term smoking, alcohol
consumption and HPV infection [44, 45, 46]. In recent years, the role of HPV has
become increasingly prominent [46]. In this study, the evaluated clinical
variables fell into three main categories: basic (gender, age, and subsite),
etiological (smoking, drinking, and HPV infection), and
tumor progression-associated (survival status, grade, tumor (T) stage, node (N)
stage, metastasis (M) stage, and TNM stage). People over the age of 65 are more
susceptible to infection [47]. Thus, we divided the patients into two groups: old
(
We assigned HNSC patients to the TPSI-high and TPSI-low groups based on the median value of TPSI and then investigated whether there was a difference between both groups for each factor. For tumor progression, only survival status showed a correlation with TPSI levels. Fewer deaths were observed in the high-TPSI group than in the low-TPSI group (Fig. 6A). From these data, we identified the prognostic performance of TPSI in HNSC. However, there was no significant difference in overall survival (OS) between TPSI-high and TPSI-low groups (Fig. 6C). Given the correlation of ACE2 or TMPRSS2 with the progression of some tumors [48, 49], we determined the prognostic role of each gene. The results showed that neither ACE2 nor TMPRSS2 expression appeared to influence the OS of HNSC patients (Supplementary Fig. 1). The two groups did present different proportions of subsites and HPV+ rates (Fig. 6B). Further comparison of TPSI distribution among the three subsites showed significantly higher TPSI levels in LH and OP than in OC. Because of this finding, the subsites were classified into two types, OC and others (LH and OP), for the subsequent analysis of variable importance. We also found that TPSI levels had an HPV+ subtype preference (Fig. 6D).
 Fig. 6.
Fig. 6.Clinical factors and gene mutations related to TPSI in TCGA_HNSC. (A) Tumor progression-associated characteristics of HNSC patients by TPSI. (B) Basic and etiological characteristics of HNSC patients by TPSI. (C) Kaplan-Meier curves for OS. (D) Distribution of TPSI scores in HNSC patients stratified by subsite and HPV status. (E) Gene mutations significantly associated with TPSI levels. (F) Distribution of TPSI levels in HNSCs with mutant NSD1, TP53, or CASP8 and their wild-type counterpart. wt, wild type; mt, mutation type. (G) Lasso regression reflecting variable importance.
We subsequently analyzed the possible relationship between TPSI levels and gene
mutations in HNSC from a genomic standpoint. To screen which gene mutations were
most significantly associated with the TPSI score, we filtered out genes with a
mutation rate of less than 10% and then performed the independence test. As
presented in Fig. 6E, NSD1, TP53, and CASP8 mutations were the most significant
mutations correlated with TPSI levels (p
Based on above observations, we sought to identify the potential key features shaping varying TPSI levels in HNSCs. Samples with missing values were excluded from the TCGA_HNSC dataset, and 487 HNSCs were included in this additional analysis. Lasso regression was conducted to measure the relative importance of two clinical variables (subsite and HPV status) and three mutational variables (NSD1, TP53, and CASP8 mutations) on influencing TPSI levels. According to the changing trajectory of the coefficient of each independent variable, the order of importance was ranked as follows: subsite, HPV status, NSD1 mutations, CASP8 mutations, TP53 mutations (Fig. 6G). Among the five features, the top three (subsite, HPV status, NSD1 mutations) were considered key factors contributing to HNSC TPSI levels, with the site of HNSC onset being the most important. The effects of these three key factors on TPSI levels were further verified by both TCGA internal validation sets (Supplementary Fig. 2).
SARS-CoV-2 is highly contagious and transmitted from person to person mainly through respiratory droplets [2]. The head and neck play a key role in the transmission of SARS-CoV-2. The head and neck include the oral cavity, oropharynx, laryngopharynx and other sites that communicate with the external environment. They may also serve as a gateway to infection due to the widespread expression of ACE2 and TMPRSS2 [50, 51, 52]. Moreover, SARS-CoV-2 infection has been confirmed in the oral cavity [25]. During the COVID-19 pandemic, many aspects of the surgical pattern and nursing care of patients with HNSC, which accounts for around 90% of all head and neck cancers were altered [53, 54]. Therefore, exploring the susceptibility to SARS-CoV-2 in HNSC was particularly important. In this study, we designed a quantitative method for measuring the potential for SARS-CoV-2 infection (“TPSI”) based on ACE2 and TMPRSS2 transcript levels, and performed a pan-cancer analysis of TPSI levels across 11 tumor types and the corresponding normal tissues. Furthermore, we investigated the factors that could influence TPSI levels in HNSC and attempted to identify the key contributors among them.
The lungs are the organs most affected by SARS-CoV-2 infection [55]. However, we observed that although the lungs were rich in ACE2 and TMPRSS2, they were not the richest among the normal organs, consistent with previous studies [56]. Accordingly, the expression of viral entry-related genes could not simply be used to compare different infection risks among different tissue types, and the specific location should also be considered. Organs such as the lungs, which are in direct contact with the outside, are more likely to come in contact with the virus and become targets for invasion. Our results showed that TPSI could reveal the internal infection potential of tissue from the level of gene expression. Thus, we recommend using TPSI in specific types of tissues rather than across different types to compare the susceptibility to SARS-CoV-2 infection or to identify predisposing factors. Pan-cancer analysis of TPSI showed that almost all (10/11) solid tumors presented lower integrated levels of ACE2 and TMPRSS2 than normal tissues, suggesting that tumor tissues are less susceptible to SARS-CoV-2 invasion. However, cancer patients are more likely to have higher morbidity and mortality of COVID-19 than the general population [8], mainly because malignancy and anticancer therapy result in immunosuppression [57]. Accordingly, the cancerous tissues are less prone to become targets of SARS-CoV-2 than the corresponding normal tissues for cancer patients when infected with this virus.
In our evaluation of HNSC, we validated lower HNSC TPSI levels in two public datasets and one of our own. Furthermore, in both the TCGA_HNSC and GSE41613 datasets, the ACE2 and TMPRSS2 expression levels were positively correlated with TPSI, and functional analysis of the TPSI-related genes showed significant enrichment of viral entry-related processes. Thus, TPSI exhibited a robust correlation with virus invading into host cells. Our results also indicated the accuracy of the TPSI signature in HNSC. Consistent with the difference in TPSI, other coronavirus entry-related host factors, translation of replicase and assembly of the replication transcription complex were more active in normal tissues than in HNSCs, suggesting that HNSC tissues are less likely to be infected with SARS-CoV-2 from the perspective of viral entry and virus replication.
We further found that the initial infiltration of two kinds of inflammatory
cells (i.e., eosinophils and Th17) increased with higher TPSI levels. Th17 can
secret interleukin (IL)-17, promoting the production of pro-inflammatory
cytokines (e.g., IL-6 and tumor necrosis factor-
Previous studies demonstrated that ACE2 exerts antitumor effects by inhibiting tumor angiogenesis [49] and promoting tumor immune infiltration [60]. TMPRSS2 is highly prostate specific due to androgen receptor regulation, which is beneficial for selective activation of EMT signaling, facilitating the metastatic process [48, 61]. However, we found that ACE2 or TMPRSS2 expression had no prognostic significance in HNSC. Moreover, TPSI, which represents their average levels, was neither related to OS nor the clinicopathologic features of HNSC. Thus, HNSC progression might not affect its susceptibility to SARS-CoV-2 infection. We also found no evident relationships between TPSI levels with gender, age, and smoking; the TPSI levels were lower in oral squamous cell carcinoma (OSCC) and HPV-HNSC.
In the analysis of gene mutations, we observed that HNSCs with mutant TP53 or CASP8 were associated with lower TPSI levels, while those with mutant NSD1 HNSCs were associated with higher TPSI levels in TCGA_HNSC. The results indicated that TP53 or CASP8 mutations might reduce the expression of ACE2, TMPRSS2, or both of these genes. In contrast, NSD1 mutations might increase the expression of one or both of these genes. Mutations in the TP53 tumor suppressor gene are the most frequently detected genetic alterations (about 70–80%) reported in HNSC [62]. Depletion of mutant p53 proteins was found to increase TMPRSS2 expression in HNSC cell lines [22], which supports our hypothesis. CASP8 is a protease involved in the extrinsic apoptotic pathway and also a negative modulator of programmed cell necrosis [63]. Mutant CASP8 HNSCs had distinctive characteristics of genes associated with inflammation and the immune response and were rich in immune cell infiltration [64]. It has been reported that inactivating mutations of NSD1 define an HNSC intrinsic subtype with significant DNA hypomethylation [65]. The “NSD1 subtypes” present an immunologically cold phenotype characterized by low-infiltrated tumor-associated leukocytes [66]. Although the gene mutations we evaluated were closely related to TPSI in the entire TCGA dataset, further lasso regression revealed that NSD1 mutations, together with lesion site and HPV status, were relatively important factors impacting TPSI levels. This finding was verified with two internal validation sets. Taken together, our data indicated that HNSCs arising outside the oral cavity, HPV+ HNSCs, or HNSCs with NSD1 mutations were more prone to SARS-CoV-2 infection.
In summary, we developed a quantitative measure called TPSI to facilitate the comparison of the integrated ACE2 and TMPRSS2 expression levels among same-type tissues or organs and to study the potential influencing factors for SARS-CoV-2 infection. We provide evidence that cancer tissues were generally less susceptible to SARS-CoV-2 infection than the corresponding normal tissues in terms of viral invasion due to the generally lower TPSI levels. Thus, it is reasonable to think that the high susceptibility of cancer patients to SARS-CoV-2 infection is caused by other factors, such as immunosuppression rather than viral entry-related genes. For HNSC, tumors occurring outside the oral cavity, positive for HPV, or containing NSD1 mutations are key factors increasing the average ACE2 and TMPRSS2 expression. These data could be used to evaluate the infection risk of cancerous lesions of HNSC patients complicated with COVID-19 and take preventive measures accordingly.
TYZ designed the experiments and drafted the manuscript; PPY and TTH contributed to data acquisition and analysis; KGZ and YMJ conducted the experiments; SJW, LLJ and BHZ critically revised the manuscript; XWZ and XY contributed to conception, design, and critically revised the manuscript.
The study was approved by the Ethics Committee of the School and Hospital of Stomatology, China Medical University (approval No. [2021] 07). Informed consent was obtained from all participants.
Thanks to all the peer reviewers for their opinions and suggestions.
Funding for this study was supported by National Natural Science Foundation of China (82071151 to B.H.Z.; 81700977 to X.Y.; 81500858 to X.W.Z.).
The authors declare no conflict of interest.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019; HNSC, head and neck squamous cell carcinoma; ADAM17, a disintegrin and metalloprotease 17; CTSL, cathepsin L; HPV, human papillomaviruses; NSD1, nuclear receptor binding SET domain protein 1; WGCNA, weighted gene co-expression network analysis; ssGSEA, single sample gene set enrichment analysis; TCGA, The Cancer Genome Atlas; Th17, T helper 17.
