† These authors contributed equally.
Background: JumonjiC (JmjC) domain-containing protein 5 (JMJD5) plays an important part in cancer metabolism. However, the prognostic value of JMJD5 in most human cancers is unknown yet. We aimed to examine the expression level and prognostic value of JMJD5, immune cell infiltration in cancer patients, and simultaneously to examine the correlations among them. Materials and methods: The mRNA and protein expression of JMJD5 were analyzed through online Tumor Immune Estimation Resource (TIMER) or immunohistochemistry (IHC) of tissue microarray sections (TMAs) in cancer versus normal tissues. The Kaplan–Meier Plotter databases were used to assess the prognostic values. The connection between the expression of JMJD5 and the abundances of six infiltrating immune cells were explored by TIMER in breast cancer (BRCA), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD) and stomach adenocarcinoma (STAD). We used the Cox proportional hazards model to investigate the correlations among clinical outcome, the abundance of immune cell infiltration and JMJD5 expression. Results: We found that the JMJD5 expression was obviously lower in BRCA, LIHC and lung cancer (LUC) but higher in STAD than in normal tissues. High expression of JMJD5 had a better prognosis only in BRCA, LIHC and LUC but a worse prognosis in STAD. The expression of JMJD5 has a significant connection with the abundance of six kind of infiltrating immune cells. The expression of JMJD5 plus the number of immune-infiltrating B cells or macrophages may jointly serve as a prognostic marker in the above four cancers. Conclusion: We provided novel evidence of JMJD5 as an essential prognostic biomarker and perspective therapeutic target in BRCA, LUAD, LIHC and STAD.
Proteins containing JmjC domains have been found as novel demethylase signature motifs contributing to variety of human cancers by means of epigenetic remodeling [1, 2]. It has been predicted that proteins containing the JmjC domain are metalloproteinases folded with copper proteins and candidate enzymes for regulating chromatin remodeling [3]. In addition to histone demethylase activity, some members of the JmjC family, such as JMJD5 and JMJD6, also have protein hydroxylase and RNA hydrogenase activities [4]. In addition to histone modifications, substrates of the JmjC protein family also include many other functional proteins, such as transcription factors, signal molecules and shear-related proteins, all of which are involved in physiological and pathological processes, such as oxidative stress and cell development [5, 6]. Further studies have shown that dysregulation of JmjC family members, e.g., JMJD5, JMJD6, JMJD2A and so on, leads to abnormal growth of embryos or causes tumor cell proliferation and migration.
The protein family which contains JmjC domain has more than 30 members, all of which have the same JmjC domain, which catalyzes the demethylation of mono-, di- or trimethylated lysines [7]. JMJD5 (also called KDM8) is one of the JmjC domain-containing protein family. Y.E. Chin and colleagues reported that JMJD5 is a cathepsin L-type protease that regulates the hydrolysis and cleavage of histone H3 N-tail protein in the stress situation, resulting in a DNA damage response [8]. JMJD5 cleaves only Kme1 H3 peptides, with little or even no cleavage function to dimethyl-lysine (Kme2) or trimethyl-lysine (Kme3), indicating that H3 N-tail cleavage plays a role in mediating gene expression [8]. Another study has shown that JMJD5 may be crucial to cell cycle regulation and that JMJD5 promotes cyclin A1 expression by affecting histone demethylation (H3K36) at the CDKN1A gene locus, further accelerating the G2/M cell cycle [9]. Knockout of JMJD5 in mice leads to embryonic lethality, suggesting that JMJD5 plays a crucial role in mammalian embryogenesis [10].
Studies of a protein similar to JMJD5 in mice have indicated a potential role for this protein as a tumor suppressor. The interaction of JMJD5 and p53 can negatively regulate p53 function during the processing of cell proliferation and cycle in human lung cancer [11]. JMJD5 is upregulated under hypoxia and subsequently plays a key part in hypoxia-induced cell proliferation and tumor metabolism in breast cancer cells [12]. Additionally, Hsing-Jien Kung and colleagues reported that JMJD5 is a suitable therapeutic target for the castration resistance and metabolic adaptation of prostate cancer cells [13]. JMJD5 is shown as a tumor suppressor function in human liver cancer pathogenesis, and JMJD5silencing can promote LIHC cell proliferation through downregulating CDKN1A transcription [14]. In contrast, JMJD5 is instead a lurking oncogene in the development of colon cancer [15].
Based on the abovementioned findings, the expression of JMJD5 is different in distinct human cancers. However, the detailed expression level, immune cell infiltration and prognostic value of JMJD5 in most human cancers are still unknown. We sought to examine the expression and prognostic value of JMJD5 as well as the connection between the expression of JMJD5 and the infiltration of immune cells in human tumors.
JMJD5 mRNA expression in various types of tumor tissues was examined by the TIMER [16, 17, 18] database (http://timer.cistrome.org/). The differential mRNA expression between tumor and normal tissues for JMJD5 has been detected using the “Gene-DE” module in TIMER across The Cancer Genome Atlas (TCGA) tumor resources. JMJD5 mRNA expression was displayed using box plots, showing the median, spread and outliers by RNA-Seq normalized by transcript per million (TPM) across normal and cancerous tissues.
All these cancer patients’ samples were obtained from Huaihe Hospital of Henan University. The present research has been approved by the Ethics Committee of Huaihe Hospital of Henan University, under the condition of written consent by each patient. All cases were diagnosed histologically by following the World Health Organization classification. All tissues were fixed in 4% buffered formaldehyde and then paraffin embedded to construct TMAs. Eight separate TMAs were generated, containing 14 different types of cancers (Table 1). The detailed IHC protocol has been previously published [19]. The following antibodies were used: rabbit anti-human JMJD5 polyclonal antibody (1:250, Abcam #28883, USA) and HRP-Polymer anti-Rabbit IHC Kit (Maixin, Fuzhou, China). Stained sections were scanned using a ScanScope T2 automated slide scanner (Aperio Technologies, Vista, CA, USA). The IHC staining was independently evaluated by two authors without knowledge of the clinicopathological information. The quantification of IHC was transformed to parameters which give the mean optical density measured using Image-Pro Plus 2.0 (Media Cybernetics, USA), the software determined the final date through optical density cumulative value divided by the target distribution area.
| TCGA Abbr. | Organ | Cancer type | T (no.) | N (no.) |
| BLCA | Bladder | Urothelial carcinoma | 20 | 20 |
| BRCA | Breast | Invasive ductal carcinoma | 20 | 20 |
| CESC | Cervix | Adenocarcinoma | 20 | 20 |
| Squamous cell carcinoma | 20 | 20 | ||
| CHOL | Biliary tract | Cholangiocarcinoma | 20 | 20 |
| COAD | Colon | Adenocarcinoma | 20 | 20 |
| KIRC | Kidney | Renal clear cell carcinoma | 20 | 20 |
| LIHC | Liver | Hepatocellular carcinoma | 20 | 20 |
| LUAD | Lung | Adenocarcinoma | 20 | 20 |
| LUSC | Squamous cell carcinoma | 20 | 20 | |
| LULCC | Large cell carcinoma | 20 | 20 | |
| OV | Ovary | Serous adenocarcinoma | 20 | 20 |
| PAAD | Pancreas | Invasive ductal carcinoma | 20 | 20 |
| PRAD | Prostate | Adenocarcinoma | 20 | 20 |
| STAD | Stomach | Tubular adenocarcinoma | 20 | 20 |
| UCEC | Uterus | Endometrioid adenocarcinoma | 20 | 20 |
The effect of JMJD5 on relapse-free survival (RFS) in the above significantly expressed cancers were investigated by using Kaplan–Meier plotter [20] (www.kmplot.com). Survival analyses were performed to generate Kaplan-Meier plots. Taking 95% confidence intervals (CIs) as hazard ratios (HRs), we obtained log-rank p values.
In order to ascertain the correlations between JMJD5 expression and six
tumor-infiltrating immune cells (B cells, CD4
Patients with BRCA, LIHC, LUAD, LUSC and STAD were divided into four groups as follows: (1) low JMJD5 expression + low tumor-infiltrating immune cells; (2) low JMJD5 expression + high tumor-infiltrating immune cells; (3) high JMJD5 expression + low tumor-infiltrating immune cells; and (4) high JMJD5 expression + high tumor-infiltrating immune cells. A Cox proportional hazards model was used to draw Kaplan–Meier plots for JMJD5 expression and immune infiltrates to visualize the survival differences. The expression of JMJD5 and six immune infiltrates was divided into low and high levels by 50%. p values of the log-rank test for comparing survival curves of four groups (2 vs. 1 and 4 vs. 3) are shown in each plot.
The data were analyzed with GraphPad Prism 5 and presented as the means
We investigated the JMJD5 mRNA expression in various cancers using
TIMER. Our results presented that the JMJD5 mRNA expression was
significantly higher in PRAD (**) and STAD (***) but lower in BRCA (*), CHOL
(***), colon adenocarcinoma (COAD) (***), kidney renal clear cell carcinoma
(KIRC) (*), LIHC (***), LUAD (
To confirm JMJD5 protein expression and estimate its clinical significance in cancers, we investigated the JMJD5 protein expression in TMAs using IHC. It is now clear the JMJD5 protein expression was significantly higher in bladder urothelial carcinoma (BLCA) (*), cervical squamous cell carcinoma (CESC) (*), ovarian cancer (OV) (*), pancreatic invasive ductal carcinoma (PAAD) (*), PRAD (**) and STAD (***) compared to the respective normal tissues (Fig. 1B) but was significantly lower in BRCA (*), CHOL (*), KIRC (**), LIHC (*), LUAD (**) and LUSC (*) compared to the respective normal tissues (Fig. 1C). The JMJD5 protein level was higher in COAD than in the respective normal tissue, but there was no statistically significant difference (Fig. 1B). The JMJD5 protein level was lower in UCEC compared to the respective normal tissue, but there was no statistically significant difference (Fig. 1C). IHC staining showed that JMJD5 was localized in different parts of tumor cells, including nuclei, cytoplasm or both, as follows: only in nuclei in BRCA and LUAD (Fig. 1C); only in cytoplasm in COAD, OV, PAAD, PRAD, STAD, CHOL, KIRC, LIHC, LUSC and UCEC (Fig. 1B,C); and in both nuclei and cytoplasm in BLCA and CESC (Fig. 1B). Additionally, we found that the protein expression and localization of JMJD5 varied according to the different pathological types of tumors. For example, the protein level of JMJD5 was lower in LUC (LUAD, LUSC and large cell carcinoma (LULCC)) compared to the normal tissues; however, there was an obvious difference in LUAD (**) and LUSC (*) (Fig. 1C) but no obvious difference in LULCC (data not shown). JMJD5 protein expression was observed only in nuclei in LUAD (Fig. 1C), only in the cytoplasm in LUSC (Fig. 1C) and in both nuclei and cytoplasm in LULCC (data not shown).
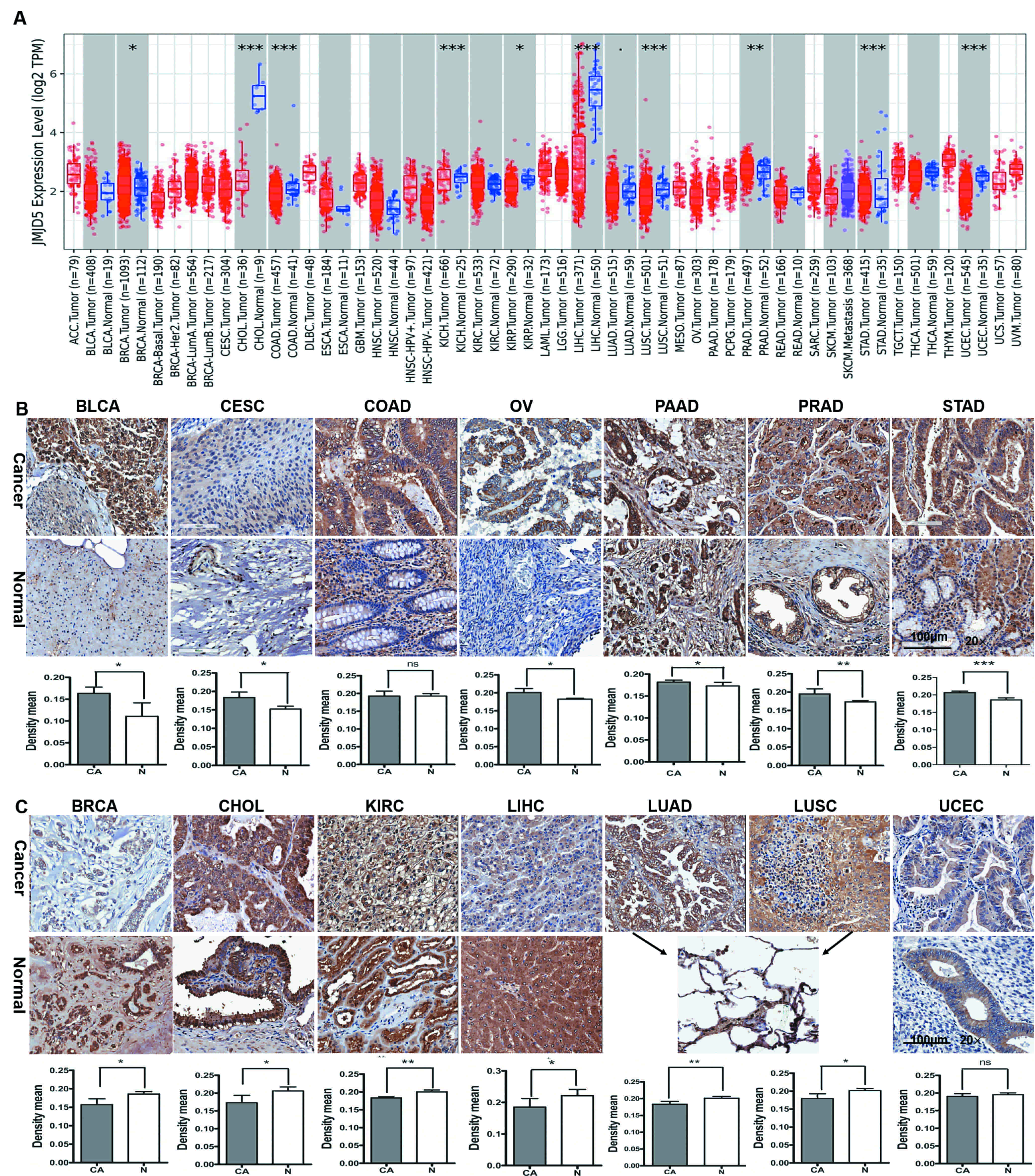 Fig. 1.
Fig. 1.Human mRNA and protein expression of JMJD5 in various
tumor tissues compared to normal tissues. (A) Box plots showing the
distributions (median, spread and outliers) of the JMJD5 mRNA levels
(log2 TPM) by RNA-seq data as displayed in gray columns when normal data were
available. The number of samples is shown at the bottom. p value
significance is indicated as follows: 0
We next determined whether the JMJD5 expression has any effect on the
prognosis of cancer patients. Among the cancers (BLCA, BRCA, CESC, OV, PAAD,
PRAD, STAD, CHOL, KIRC, LIHC, LUAD and LUSC) with significantly different
expression of JMJD5, we identified JMJD5 as a prognostic marker
in BRCA (n = 3955, RFS: HR = 0.75, 95% CI from 0.67 to 0.83, log-rank p =
1.4
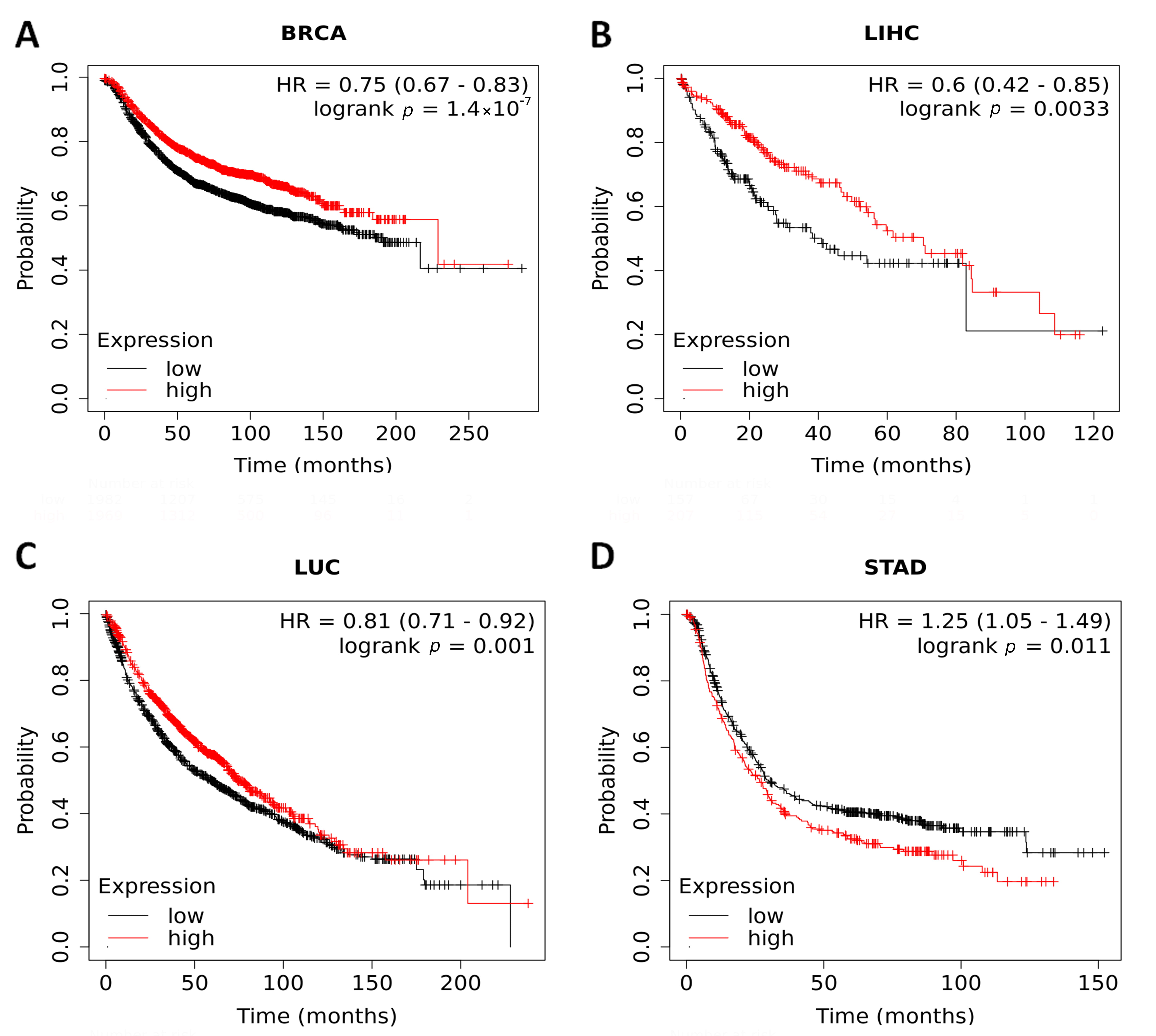 Fig. 2.
Fig. 2.Kaplan–Meier survival curves comparing the low and high
expression of JMJD5 in BRCA, LIHC, LUC and STAD patients. (A) RFS of
BRCA. (B) RFS of LIHC. (C) RFS of LUC. (D) RFS of STAD. The red curve indicates
patients with high JMJD5 expression, while the black curve indicates
patients with low JMJD5 expression. p
We focused on and analyzed the correlation between JMJD5 expression and
the quantity of six infiltrating-immune cells (B cells, CD4
 Fig. 3.
Fig. 3.Correlation of JMJD5 expression with six
tumor-infiltrating immune cells in BRCA, LIHC, LUAD, LUSC and STAD. (A) In BRCA,
JMJD5 expression significantly negatively correlated with tumor purity
and neutrophil infiltration and positively correlated with the infiltration of B
cells, CD8
The Kaplan–Meier plots showed that the survival rate of low JMJD5 expression patients with higher B cell tumor infiltration were much better than those with lower B cell tumor infiltration in BRCA (n = 1100, OS: HR = 0.658, p = 0.0487) (Fig. 4A; 2 vs. 1) and LUAD (n = 515, OS: HR = 0.67, p = 0.039) (Fig. 4B; 2 vs. 1). The survival rate of high JMJD5 patients with lower macrophage tumor infiltration were much better than those with higher macrophage infiltration in LIHC (n = 371, OS: HR = 1.72, p = 0.0158) (Fig. 4C; 4 vs. 3). The survival rate of low JMJD5 expression patients with lower macrophage infiltration is much better than those with higher macrophage infiltration in STAD (n = 415, OS: HR = 2.26, p = 0.00035) (Fig. 4D; 2 vs. 1). In summary, the combination of JMJD5 expression with either B cell tumor infiltration or macrophage tumor infiltration may serve as a new tumor prognostic marker.
 Fig. 4.
Fig. 4.Kaplan–Meier plots for expression level and immune cell infiltrates to visualize the survival differences in BRCA, LUAD, LIHC and STAD. (A and B) The survival rate of low JMJD5 expression patients with higher B cell tumor infiltration were much better than those with lower B cell tumor infiltration in BRCA (A) and LUAD (B). (C) LIHC patients with high expression of JMJD5 had an improved survival rate with lower macrophage tumor infiltration compared to those with higher macrophage tumor infiltration. (D) The survival rate of low JMJD5 expression patients with lower macrophage infiltration is much better than those with higher macrophage infiltration in STAD.
The JmjC domain-containing protein family contains more than 30 members, and many members are aberrantly expressed or dysregulated in many kinds of human cancers and regulate the proliferation and invasion of tumor cells [21, 22]. For example, the aberrant expression of some family members, such as PHF8, KDM3B and JMJD2A, promotes the proliferation and metastasis of tumor cells in PRAD and BRCA [6, 23]. JMJD5 belongs to the JmjC domain-containing protein family, but the expression level, prognostic value and the correlation of tumor immune infiltration in most human cancers are still unclear. From our study, we reported the protein expression of JMJD5 in almost all human cancers for the first time, and we found that JMJD5 was overexpressed in STAD and that high expression of JMJD5 indicated poor survival. In contrast, low expression levels of JMJD5 were found in BRCA, LIHC and LUC, and low JMJD5 expression was yielded a poor outcome. Therefore, JMJD5 is not only a potential prognostic biomarker but may also be a therapeutic target for BRCA, LIHC, LUC and STAD.
JMJD5 is a nuclear protein which mostly move between cell nucleus and cytoplasm [4, 24]. JMJD5 has many enzyme activities, including H3K36me2 demethylation activity, C3 arginine hydroxylation activity, endo-/exopeptidase activity at arginine-methylated histones and endopeptidase activity at lysine-methylated histones, and this function may closely correlate with various human diseases, such as tumors, diabetes and so on, through epigenetic regulation. JMJD5 is involved in different physiological and pathological processes. Recent molecular mechanism study showed that JMJD5 activates or suppresses gene expression at the transcriptional and posttranslational levels. Thus, JMJD5 may play a role in pro-cancer or anticancer activity depending on context.
Recent research has reported that immune cells present in the microenvironment of tumor either inhibit or support the growth and development of tumors [25]. Tumor-infiltrating immune cells contain those mediating adaptive immunity, T lymphocytes, dendritic cells and occasional B cells as well as effectors of innate immunity, macrophages, polymorph nuclear leukocytes and rare natural killer cells [26]. Recently, studies have shown that the B cells existing in human tumors is associated with a promising response to immunotherapy [27, 28, 29]. Furthermore, macrophages existing in tumors named tumor-associated macrophages are reprogrammed to suppress lymphocyte functions by releasing of inhibitory cytokines [30, 31]. To date, JmjC domain-containing protein family has been hardly reported in terms of in-depth study in immuno-oncology. There is no report on the relationship between JMJD5and immune cell infiltration. Our results revealed that the mRNA expression of JMJD5 may reflect immune cell infiltration in BRCA, LIHC, LUAD, LUSC and STAD and that infiltration by high B cells is a key discriminative feature of patients with low JMJD5 in BRCA and LUAD with improved survival. Additionally, we found that the number of immune-infiltrating macrophages combined with JMJD5 expression may serve as a prognostic marker for LIHC and STAD. This finding may have broad applications in tumor targeting therapy and immunotherapy.
In summary, we provided novel evidence of JMJD5 as an essential prognostic biomarker in BRCA, LIHC, LUAD and STAD. Our future studies will aim to determine how to regulate the expression of JMJD5 and tumor-infiltrating B cells or macrophages in BRCA, LUAD, LIHC and STAD, which may be a promising therapeutic approach in tumor treatment.
ZS designed the study; HL, QL, HJ, JZ, HZ and XM performed the research; ZS, HL and QL analyzed the data and wrote the paper; WS, HL and QL revised the paper; LW and RD contributed reagents and materials. All authors reviewed and approved the final manuscript.
All samples were obtained from patients with cancers who had surgery in Huaihe Hospital of Henan University, and written informed consent was obtained from each patient. This study was approved by ethics committee of Huaihe Hospital of Henan University, code: HUMOR2020-112.
Thanks to all the peer reviewers for their opinions and suggestions.
This research was supported by the Henan Science and Technology Planning Project (grant no. CX0001F0010344).
The authors declare no conflict of interest.
BLCA, Bladder urothelial carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervix squamous cell carcinoma; CHOL, Cholangiocarcinoma; COAD, Colon adenocarcinoma; HR, Hazard ratio; IHC, Immunohistochemistry; JmjC, JumonjiC; JMJD5, JumonjiC domain-containing protein 5; KIRC, Kidney renal clear cell carcinoma; LIHC, Liver hepatocellular carcinoma; LUC, Lung cancer; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; OS, Overall survival; OV, Ovarian cancer; PAAD, Pancreas invasive ductal carcinoma; PRAD, Prostate adenocarcinoma; RFS, Relapse-free survival; STAD, Stomach adenocarcinoma; TCGA, The Cancer Genome Atlas; TMA, Tissue microarray; 95% CIs, 95% confidence intervals; UCEC, Uterine corpus endometrial carcinoma.
