Send correspondence to: Ankur Kaushal, Amity University, Haryana-122413, India, 2Shoolini University, Solan- 173229, India, Tel: 91-9882546292, Fax: 01792-308000, E-mail: ankur.biotech85@gmail.com
Leptospirosis can be found in virtually all tropical and temperate areas of the world and is presumed to be the widely spread zoonotic infection in the world. Because of the variety of clinical symptoms seen in the symptomatic cases, leptospirosis at its onset is often misdiagnosed as aseptic meningitis, influenza, hepatic disease or fever (pyrexia) of unknown origin. The disease has been widely spread, ranging from subclinical infection to a severe syndrome of multiorgan infection with high mortality. It is an occupational hazard for people who work outdoors or with animals, such as rice and sugar-cane field workers, farmers, sewer workers, veterinarians, dairy workers, and military personnel. Various diagnostic methods have been developed for the diagnosis of leptospirosis that includes direct examination; serology and molecular based techniques, but have various shortcomings, so there is a need to develop an effective surveillance system to monitor the trends of disease to control this life-threatening zoonosis. Now a day’s biosensor based technology becomes an excellent platform in the field of diagnostics due to their better sensitivity and specificity. So different types of biosensors such as enzyme-based, tissue-based, immunosensor, DNA biosensors, thermal and piezoelectric biosensors have been discussed here to highlight their indispensable applications in different fields. In this review, we will examine the current utilization of functionalized detection methods with other synthetic mixes for the development of biosensor prompting to the location of particular analytes with low discovery cut-off and quick reaction.
The advancement in the field of medicine has led to the gain of overall health benefits to the population. The diseases which were thought to be untreatable and were responsible for the mass evacuation some decades ago are now very easy to detect and get treated. Many causative microorganisms have been identified and the ways to diagnose their treatment have been implemented for various diseases. However, as advancements have been increased, microorganisms challenge the researchers by developing resistant mechanisms and the emergence of new strains. At times, some microbes are not intended to infect the human population but they do by chance and thus are responsible for the death of individuals. HIV is one of the examples of such a microbe whose causative organism were monkeys but by chance, it got infected to humans and to date human population is tackling a way to get rid of this particular virus. Similarly, leptospirosis is one of the diseases which accidentally got into the human population via rodents and possess a very big threat. Leptospirosis is the zoonotic disease caused by a gram-negative spirochete Leptospira (1). The disease is characterized by the identification of thin, coiled, aerobic bacteria in the body fluids of the infected people (2). A wide range of animals acts as a host for leptospira including fishes, amphibians, birds, reptiles, and mammals. Exposure to environmental sources, such as animal urine, contaminated water/soil, or infected animal tissue remains the biggest possibility of humans acquiring the disease (3).
The bacteria can also infect people by the consumption of food contaminated with urine or inhalation through aerosols. The people associated with farming (in contact with water), rodent catchers are more likely to get this disease (4). Leptospira is transmitted to people usually through direct contact of skin with contaminated soil, water and flora with the urine of infected animals. The infection can also be transmitted by direct contact with infected animal’s urine, tissues and even by inhalation of contaminated fluid. The bacterium enters inside the system through cuts in the skin, mucous membranes, or conjunctivae. The bacterium is known to possess the ability to even enter through the intact skin. The transmission through ingestion of food materials contaminated with urine of infected rats is occasional (5). Different studies have been carried out to understand the mechanism of leptospira pathogenesis, which is still not well defined. Recent studies reported the role of leptospiral extracellular proteases in its pathogenesis, as it shows proteolytic activity against host proteoglycans and plasma proteins, most likely with the participation of metalloproteases. There are various receptors that start the host immune response after leptospira infection. Toll like receptors are the transmembrane protein receptors that play a major role in primary immune response. Bacterial proteases hydrolyze diverse proteinaceous substrates of the host and play a crucial role in colonization and spreading, allowing evasion of innate immune responses and contributing to disruption of tissue integrity (6). In this regards body generates a significant immune response against the surface lipopolysaccharide (LPS) (7). After this response various circulating cytokines and chemokines have been induced as a secondary response of the immune system. Despite of the severity of the leptospirosis, the molecular pathogenesis of the disease is still not completely understood. Loa22 surface lipoprotein is the main virulence factor responsible for disease pathogenesis (8). Thus, it becomes the need of the hour to devise a novel technique for the early diagnosis of the disease to provide timely treatment to the patient. This review discusses the recent advances which have been taken in the diagnosis of the disease and their benefit over the traditional standard diagnosis kits in terms of cost, sensitivity, specificity and time.
Leptospira is the genus belonging to the order spirochetes, family leptospiracea with the body diameter and length of 0.1 µm and 6-20 µm respectively is comprised of several pathogenic and saprophytic species (9). The genus is characterized by the morphological features such as flexibility, spirally shaped (distinctive hooked ends) and the presence of two periplasmic polar inserted internal flagella for motility (10). The cell wall of the bacteria is composed of lipopolysaccharide (LPS) which is homologous to other gram-negative bacteria. The data about heritability of leptospirosis is not still clear but in leptospira the variation in the structural lipopolysaccharide (sugar composition and orientation) is associated with identification of over 200 serovars of the leptospira. Leptospires are classified clinically and genotypically. Clinically, leptospires are classified into pathogenic (parasitic and infectious) and non-pathogenic strains (Saprophytic and non-infectious), whereas genotypic classification relates to the different species based on the phenotypic relationship. Overall, non-pathogenic strains are known to occur in free-living state including L. biflexa, L. meyeri, L. yanagawae, L. kmetyi, L. vanthieli, and L. wolbachii with over 60 serovars and pathogenic species are parasitic, disease-causing species including L. interrogans, L. borgpetersenii, L. santarosai, L. noguchii, L. weilli, L. kirschneri and L. alexanderi with over 250 serovars (11). However, both serological and molecular classifications of Leptospira are not directly correlated and a given serogroup is often found in several Leptospira species. For example, the serovars of the Bataviae serogroup are found in serovars of L. interrogans, Leptospira borgpetersenii, Leptospira santarosai, Leptospira kirschneri, Leptospira noguchii and Leptospira borgpetersenii. The higher genetic and serological diversity among Leptospira required identification of the circulating species/serovars in order to enhance leptospirosis prevention and control strategies (11). LPS forms the major antigen for the recognition of the bacteria by the host immune system. The genome of Leptospira is about 3.9 - 4.6 Mbp composed of two circular chromosomes although a third replicon (p74) is found in non-pathogenic strains. However, thirteen genes of p74 are known to be present in the first chromosome of the pathogenic strains (12).
The optimal growth temperature is found to be 28ºC in a nutrient-rich media which is solely composed of carbon sources such as vitamins (B1 and B12), long-chain fatty acids associated with albumin and ammonium salts (13). The multiplicative ability of the leptospires is hindered by certain environmental factors and the host unavailability. The bacterial proliferation is known to be affected by several environmental factors such as humidity, temperature, pH and the presence of an inhibitor. Both pathogenic and non-pathogenic strains are morphologically identical but can be distinguished by observing their growth in nutrient media-rich in 8-azaguanine in low temperature (8-azaguanine as an inhibitor for pathogenic strain) and by measuring the pathogenicity in animals (14). To understand the colonization of the bacteria, several animal models have been established among which hamsters and guinea pigs being the most commonly used. An intraperitoneal leptospiral load of 103-108 is required to initiate infection (LD 50 calculated for different strains) (15, 16). Factors responsible for virulence have also been identified by gene knockout and site-directed mutagenesis techniques; thereby the essential genes for virulence have also been confirmed. The genes which were found responsible included flaB, rec A (code for flagellin protein), trpE (tryptophan biosynthetic gene), hemH (heme biosynthetic gene), metX (methionine biosynthetic gene), ligB (outer surface gene), lipL32, loa22 (adhesion protein), fli Y (flagellar motor switch) and fecA, feoB (iron acquisition) (17). Investigations by pairwise alignment and phylogenetic tree construction have suggested that the presence of virulent lipL32 protein has been found in fourteen leptospira pathogenic strains with limited diversity in terms of pairwise alignment, hydrophobic groups, hydrophilic groups and number of turns. This conservation analysis led the investigators to look into the gene as a potential target antigen for leptospiral diagnosis and to evaluate its potency as a novel target virulent gene in the early diagnosis of leptospirosis (12).
The leptospiral infection is clinically termed biphasic due to the occurrence of two phases (leptospiremic and leptospiruric) simultaneously. The phases are characterized by the bacterial presence at various sites and the corresponding immune response generated. The leptospiremic phase is characterized by the penetration of the leptospira in the blood vessels, cerebrospinal fluid and the generation of specific IgM and IgG antibodies that forms the preliminary diagnosis method for the detection of leptospirosis. In the leptospiruric phase, the infection reaches to multiple organs such as lungs, liver, kidneys, heart, skin, muscles and cerebrospinal fluid in the brain resulting in several diseases associated with the acute leptospiral infection such as vasculitis in capillaries (endothelial edema, necrosis, and lymphocytic infiltration), interstitial nephritis and tubular necrosis in kidneys (tubular damage and renal failure), hepatocellular dysfunction, centrilobular necrosis and Kupfer cells proliferation in liver (jaundice), alveolar and interstitial damage in lungs (haemorrhage), cardiac lesions in heart (interstitial myocarditis) and edema, myofibril vacuolization, and vessel damage in skeletal muscles. Although the host immune system reacts by secretion of specific opsonising antibodies followed by immediate clearance causing symptomatic inflammatory reaction producing secondary end-organ injury (18). However, the bacteria reside in immunologically privileged sites (renal tubules, brain, and aqueous humour or eyes) of the surviving host for months and cause chronic infection symptoms such as recurrent uveitis and appearance of leptospires in urea (19).
The existing treatment of leptospirosis includes the manifestation of antibiotics and the development of vaccines. A range of antibiotic groups has been found useful to treat the individuals in their first phase of infection such as doxcycline, penicillin (penicillin G), ampicillin (omnipen, Polycillin), amoxicillin (Amoxil, Trimox) and erythromycin (pediazole, Ilosone) (20, 21). These antibiotics though have a significant effect on the patients but they are not recommended for long term use as well as in the later phases of the disease. The development of a vaccine has helped the high-risk workers but unfortunately, these have been made specific to only some of the serovars as developing specific vaccines to all the serovars individually is physically and economically impossible thus making people susceptible to other serovars. The generation of primary antibodies in the patient's serum is time-consuming and also the concentration is very low. So the use of antibiotics (benzylpenicillin, erythromycin and doxcycline) in the treatment of leptospirosis is the gold standard. But the treatment of the patient depends upon the severity of the infection as in case of severe infections like renal failure and Weil’s disease, hemodialysis and platelet transfusion can be done (22). Thus it becomes of utmost importance to devise a novel method of early diagnosis of the disease so that the individual could be treated effectively and timely.
Leptospirosis is known to occur both in temperate and tropical regions. However, the tropical incidences are approximately 10 times higher as compared to temperate regions (23). Although leptospirosis is an under-reported disease, Leptospirosis Burden Epidemiology Reference Group (LERG), an organization under the World Health Organization (WHO), walks upon global annual incidence of endemic and epidemic human leptospirosis. WHO uses the Global Burden of Disease (GBD) approach to report health information where Disability-Adjusted Life Year (DALY) is calculated equating to the loss of one year of a healthy life. The calculation includes both the years of life lost due to premature death as well as the years of life lost due to a disability-related to the disease under investigation (24).
To estimate the age and gender-specific incidence of leptospirosis, the Monte Carlo model was developed to obtain estimates for incidences and mortality caused by the disease. 14.77% of cases were estimated to be occurring per 100,000 people annually. Globally, the disease was found to be prevalent in the countries that lie between the equatorial line of tropic of cancer and tropic of Capricorn owing to the burden of 1,030,000 cases with 58,900 deaths annually (25). The severity of the leptospirosis disease can be understood by knowing the fact that around 10,000 cases of severe leptospirosis are hospitalized every year throughout world and has high mortality rate. Another study confirms that annually around 853,000 cases have been reported on symptomatic basis worldwide. The mortality rate is higher in tropical regions that include Asia Pacific and Latin America region (26). According to the Integrated Disease Surveillance Project (IDSP) data of 2018 in Kerala region of India there were total 1318 confirmed leptospirosis cases with 95 deaths; with a death rate percentage is around (4.0%) (27). Males age between 20 and 29 were found to be more affected by the disease according to this model although the mortality index was higher in the male's age 50-59. This model allows one to estimate the global burden of disease based on age, gender, and geographical areas to evaluate the strategies that may be required for prevention.
Later a study to demonstrate the Seroprevalence in trapped rodents (Myocastor coypu, Ondatra zybethicus, Rattus norvegicus, Rattus rattus, Oryctolagus cuniculus, and Herpetesicus javanicus) was performed. The seroprevalence was calculated based on the presence of agglutinating antibodies against the leptospires in their serum. The seroprevalence was found to be around 48% in myocastors, 50% in rats and 47% in mongooses thus indicating a strong role in the transmission of the pathogenic strain (28).
The biasness in the occurrence of the disease can be noticed by the fact that the incidences of the disease are relatively low in the developed areas in comparison to developing areas. Major outbreaks of this disease have been seen after heavy rainfall or floods as the risk of exposure to contaminated drinking water also increases. Flooding in developing South Asian countries such as Bangladesh, India, and Sri Lanka have caused overflowing of rodent-infested sewers thus, exposing inhabitants to a leptospirosis infection. A similar outbreak of leptospiral infection was witnessed across Central America after Hurricane Mitch in 1998. Apart from the environmental crisis, the disease is also associated with occupational and leisure exposure. An outbreak was witnessed among the participants of the swimming triathlon in Illinois where 12 percent of total athletes were recorded to be affected and in Borneo, Malaysia where 44 percent of the athletes reported the positive infection. The outbreaks were also witnessed in triathlon happened in Germany and Austria in 2006 and 2010 respectively. Besides the risk of acquiring the disease from the environmental means, the transfer of leptospiral infection through the placenta to the fetus is also known. A study reviewed the consequences of the fourteen pregnancies of affected mothers out of which 8 were having spontaneous abortions and four infants were born with the infection. Another similar study indicated about 50% risk of fetal abortion or death with the infected mother (29).
Leptospirosis is known to be endemic in India since the early 20th century. Significant numbers of outbreaks have occurred in the coastal regions of the states of Gujarat, Maharashtra, West Bengal, Orissa, Kerala, Tamil Nadu, Karnataka, and the Andaman Islands. Highest rates occur during October to November that coincides with the monsoon season in these parts (30- 34). In 2007 an outbreak in Karnataka was reported in which 1,516 cases were found to be affected (35). Leptospiral endemic in southern Gujarat reported 130 deaths within two months due to leptospirosis in 2011 (36). Recently, 209 cases with 12 deaths were reported from Kochi, Kerala and 16 deaths were reported from Surat and Valsad districts of Gujarat (37). These alarming reports underline the risk and severity of the disease and require better and specific diagnostic tools (38).
The diagnosis of leptospira includes three different methods i.e., general laboratory findings, indirect methods, and direct methods. All these are explained in detail in subsequent sections.
To confirm that the person may be infected by the leptospira infection, firstly, the nonspecific characteristics are assessed by assessing blood, urine, and Cerebrospinal fluid (CSF) of the suspected patients. The occurrence of leukocytosis and thrombocytopenia with elevated levels of creatinine, urea, aminotransferases, bilirubin, alkaline phosphatase in blood, occurrence of proteinuria, pyuria, microscopic hematuria, hyaline and granular casts in urine and presence of pleocytosis, xanthochromia and elevated level of CSF in suspected patients suggests that the individual might be affected by leptospira infection. But, specific microbiological assays are required to confirm the infection (39). However, the facility to undergo serological tests is available only in some laboratories with low sensitivity (40). The severity of the infection can also be verified by isolating the bacteria from the tissues such as liver, kidneys, eyes, and skin. Nevertheless, it is difficult to acquire culture. Thus, different diagnostics assays are performed for confirmation.
Direct microscopy and dark field phase contrast microscopy are employed to detect leptospira in body fluids such as blood, urine, and CSF (Figure 1). The leptospira can be visualized by their morphological appearance (thin bright actively motile rods) and the characteristic rapid spinning and jerking motility. The disadvantage of this method lies in the fact that the minimum number of Leptospira to be visualized is approximately 10 cells/ml and the positive result decreases to 90% if the infection exceeds one week. Additionally, it is very difficult to analyze the false positives and false negatives with this method of diagnosis (41). However, microscopy can be coupled with silver stains such as warthin starry stain or immunostaining with specific antibodies but it is not advantageous in early infections (42, 43).
 Figure 1
Figure 1Leptospira cells observed under a microscope.
Primary culture can be grown from urine, blood and CSF samples of the patients by growing the sample in EMJH (Ellinghausen McCullough Johnson Harris Medium) media enriched with 8-10 % v/v rabbit serum supplemented with rifampicin, neomycin, and actidone. The growth of the leptospires in the medium can be visualized by dark field microscopy and measured in the terms of the CFU/mL. The disadvantage of this type of diagnosis is that culture growth is time-consuming because of less number of circulating organisms in the blood and it becomes impossible for the current diagnosis. Additionally, the doubling time of the organism is high and due to its highly infectious nature, the Biosafety cabinet level 2 is required that becomes the limiting factor for most labs lacking this facility (44).
The rapid and direct diagnosis is enabled by direct Polymerase Chain Reaction (PCR) and its variants (nested PCR and PCR/RFLP for 16s ribosomal RNA) on specimens (culture and patient’s fluids) in both early and recuperative stages of infection (Figure 2). The advantage lies in the fact that reaction can detect leptospiral DNA in the specimen at extremely low levels, accounting for the DNA content of even less than 10 leptospires. A supporting study on 103 patients of meningitis of unknown cause was performed and results showed 39.08% positive suspects by PCR, 3.88% by ELISA & 8.74% by MAT (45). However, some studies also confirm that the sensitivity of the diagnosis increases when two of the diagnostic methods via, PCR and ELISA are performed simultaneously (46, 47).
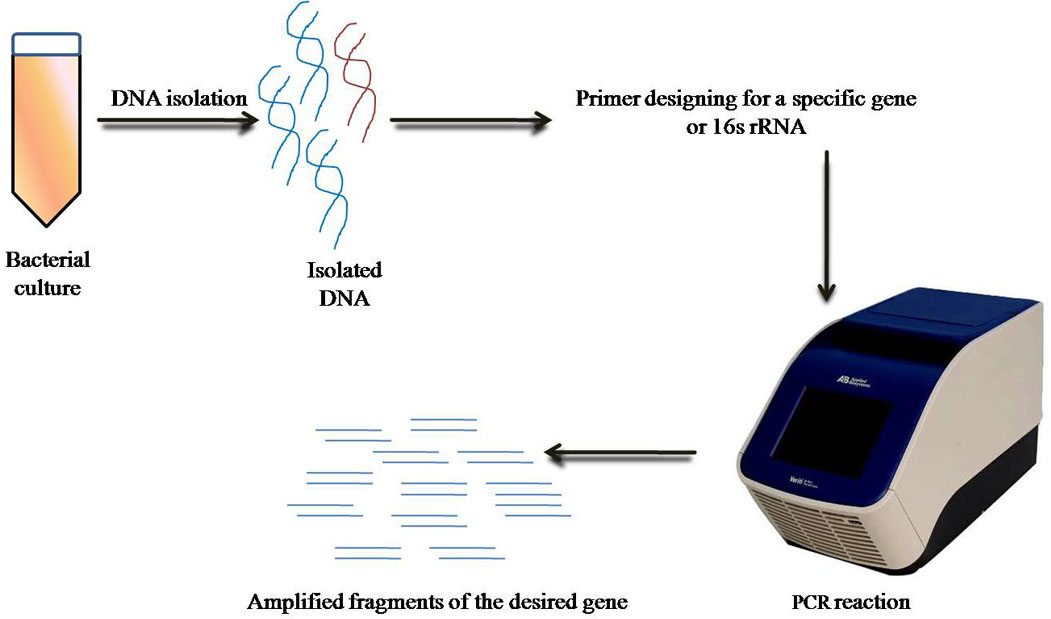 Figure 2
Figure 2Schematic representation of PCR.
Despite the advantages like increased sensitivity, PCR-based diagnosis of leptospirosis is limited by its inability to the identification of the infecting serovar (48, 49). Additionally, diagnosing the causative organism directly from the blood by PCR leads to the interference of the host's blood DNA in PCR by non-specific primer binding and the presence of ions and proteins present in blood stated that the presence of Fe+3 ion inhibits the replicating enzyme DNA polymerase (50).
Serological methods are commonly used for the diagnosis of Leptospirosis and can detect this disease after the 6th day of the disease onset. In this, the detection of the serum antibodies using Microscopic agglutination assay (gold standard), Enzyme-linked immunosorbent assay (ELISA) and Lepto dipstick assay are the most commonly used techniques.
Microscopic agglutination test (MAT) takes into account the assessment of antibodies formed in the blood of the suspected patients by their interaction with the live leptospiral antigens. In this test, the 7-12 day old leptospiral culture is made to interact with the serum antibodies of the patients in the well plates. The antigen-antibody interaction is measured in the terms of agglutination visible under a dark field microscope (Figure 3). MAT is considered as the gold standard for leptospiral diagnosis as it is a sensitive assay, but since leptospira species possess antigenic heterogeneity, a large number of serovars are required to be considered as antigens. So this test is not useful at the early stage of disease diagnosis either lack of specific antibodies in the immune system or Ab-titre is very less that leads to false-negative results (51). Moreover, if the patient is previously infected with a different serogroup, then, the diagnosis is furthermore complicated due to the anamnestic response (first antibody titer rise against the infecting serovar from previous exposure) (52). Although MAT is routinely used as a diagnostic test for leptospirosis in clinical laboratories, novel ways need to be reviewed due to a lot of serious concerns associated with this gold standard.
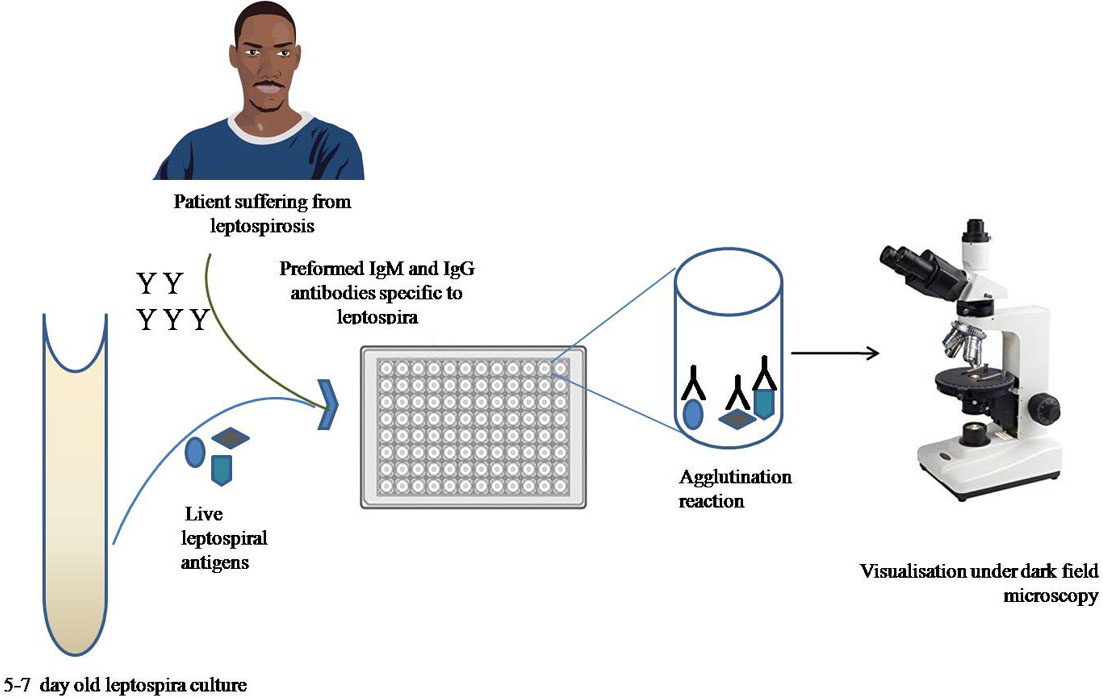 Figure 3
Figure 3Schematic representation of Microscopic agglutination assay (MAT).
ELISA also detects IgM antibodies appearing in the blood however; MAT is used as the reference test (Figure 4). The advantage of ELISA over MAT is that it does not require the use of live antigens for the diagnosis. However, this way of diagnosis is discrete as single sera testing for suspected patients is not enough for the confirmation of the disease and requires confirmatory test by testing sera of suspected patients after two weeks as well (53-55). Another limitation for the use of this diagnostic method is the time required for the formation of IgM antibodies, its appearance and detection limit in the serum (56). Moreover, the persistence limit of the IgM antibodies in the serum is very long thus the probability of occurrence of false positives and false negatives are more. An in house ELISA, variant of traditional ELISA, was developed which uses Leptospira fainei (formalin-treated or boiled) as an antigen to detect Leptospira-specific IgM antibodies in the sera of suspected patients. To indicate a statistically acceptable agreement between MAT (gold standard) and In house ELISA, a kappa value of 0.92 (excellent agreement) was observed. Such agreement between in-house ELISA and MAT suggested that the technique can be applied as a rapid screening test for the diagnosis of leptospirosis in laboratories and hospitals where MAT is not available (57).
 Figure 4
Figure 4Schematic representation of ELISA.
IHA testing is yet another way of detecting antigen-antibody (genus-specific) interaction in the serum samples of the suspected patients. The test works on the principle of passive agglutination reaction where the erythrocytes adsorb the antigen and interact with the soluble antibodies in the serum of the suspected patients. And thereafter the granular ring of an agglutinating complex in in-vitro conditions (microtitre plate) is detected for the diagnosis of leptospirosis (Figure 5) (58). It is a rapid and easily performed method compared with the MAT but has lower specificity and sensitivity. Another disadvantage of this assay is the requirement of enough amounts of the antibodies in the suspected patients which takes around eight days to form and thus has a very limited scope in the diagnosis of leptospirosis (59).
 Figure 5
Figure 5Schematic diagram of indirect Haemagglutination assay (IHA).
Apart from MAT, ELISA and IHA tests which have gained popularity in the leptospiral diagnosis over time, other tests such as lepto-dipstick Assay, immune-fluorescence Assay (IFA) and Leptospira immune-chromatography test have also been developed for the diagnosis of the disease. All these assays detect the antibodies formed in the serum after the infection of the leptospirosis (60). The accuracy, specificity, and sensitivity of these assays is confirmed by Bayesian class latent modeling in which various studies confirmed that none of the tests qualifies to be the better technique for the diagnosis of leptospirosis and thus, there is an utmost need for the development of a novel diagnostic method which can be qualified as a new gold standard for the diagnosis against the disease (61-65).
This review article proposes the biosensing method of diagnosis for leptospirosis that can overcome the disadvantages of the conventional diagnostic assays in terms of specificity, sensitivity, and limit of detection. Different biosensing techniques have already been developed for the diagnosis of various pathogenic bacteria species as discussed in the subsequent sections.
The conventional methods of detection of various bacterial diseases are time-consuming and laborious having less specificity and sensitivity. The biosensor based diagnosis can overcome the disadvantages of the conventional diagnostic assays in terms of specificity, sensitivity, and limit of detection. Some bacterial infections such as sexually transmitted infection (STI’s) however, the delay is not that critical in terms of clinical outcomes but early diagnosis is required to prevent the spread (66, 67). The early and accurate diagnosis of food borne pathogens is also required as in some cases; the deaths associated with food poisoning are a matter of concern. In addition to public health, an early warning system is also required in case of the identification of deadly microorganisms that are causing bioterrorism (Bacillus anthracis, Yersinia pestis) (68,69). That means there is a presence of thousands of microorganisms affecting us directly and indirectly, so we need to develop some instant point of care diagnostic devices having more sensitivity and specificity.
Biosensors are the devices that combine the biorecognition elements with transducers and convert an otherwise non-detectable bio-signal to a measurable readout thus confirming the biological interaction (enzyme-substrate reaction, antigen-antibody reaction, DNA-DNA interaction, DNA- protein interaction). The classification of biosensors is based on either signal transduction (electrical, chemical, mechanical and optical) or the type of bio-recognition (affinity and catalytic). The array of applications fulfilled by biosensors is very broad and recently researchers have found their applications in disease diagnostics with great specificity and sensitivity. Thus, the Diagnosis of microbial infection commonly exploits the affinity-based sensors with the use of amperometric or optical transduction for the detection of the microorganisms although, much recently, the use of impedance and fiber optics-based transduction are also in use (70).
Biosensors offer a rapid, easy and cost-effective lab-on-a-chip method of microbial detection thus monitoring and preventing microbial infections. Bacterial sensing can be performed in two ways, one requiring the bacterial pre-processing to target bacterial components such as DNA, RNA or specific intracellular proteins and other one is the development of the processing free systems that sense the whole microbe (surface epitopes, bacteriophages, polyclonal antibodies and cell wall components such as glycoprotein, lipids, and proteins). Whole-cell sensors for the detection of various gram-negative (H. influenzae, Salmonella, N. meningitides, Legionella, E. coli, N. gonorrhea, and Shigella) and gram-positive bacteria (Bacillus, Streptococcus, Clostridium, and Staphylococcus) have been developed so far. The main aim is to make new diagnostic methods to be cost-effective. Along with the advantage of cost-effectiveness, the non-processing systems let us sense a very low dose of pathogens present in the body fluids (eg, E.coli O157: H7 in food or environmental samples) (71-78). The whole microbe biosensors are undoubtedly inexpensive, small, and label-free, with little or no sample preparation but the usage of whole-cell infectious microbes requires sophisticated laboratories with the desired types of equipment and experienced technicians to handle the microbe which is not available with every laboratory. This major factor leads the researchers to approach the direction of development of a system that requires the pre-processing of the live microbe and senses the presence of internal proteins, DNA or RNA which makes it much easier to operate without the requirement of experienced personnel. Apart from the biological interaction, another component of the biosensor is the transducer which is an electrical component used to detect the bio-signal. This component can employ various principles of detection and based on this, biosensors are classified as optical, mechanical, impedimetric, amperometric, electrochemical and potentiometric biosensors.
Optical biosensors work on the basic principle of bio-recognition induced changes in the optical properties of the surface of the sensor. They are classified into two categories viz, fluorescence-based and label-free (79). The optical biosensors function by detecting and measuring a change in absorbance, luminescence or fluorescence of the biosensor surface when the bio-receptor recognizes specific analyte. The bio-recognition element comprises immobilized biomolecules (for instance enzyme or antibody) which allow for specific analyte detection. The bio-recognition is followed by the binding of a secondary reagent, for instance fluorescently labeled antibody to the captured analyte on the surface of the sensor which in turn generates a detectable optical signal, whose strength is proportional to specific binding of the analyte to its receptor. Furthermore, a portable biosensor system employing optical fibers has been used for the detection of microbes (80, 81). Fiber-optic biosensors (FOB) are optical fiber derived sensors that exploit the mechanism of optical field to detect the specific bio-receptor-analyte binding. The setup is comprised of a source of light and a detector where light passes through the optical fibers onto which the interest bio-receptor is immobilized. The specific bio-receptor-analyte binding is followed by the addition of labeling reagent which leads to a detectable change in optical signal and is further detected by the detector. Despite having advantages such as low limit of detection, and selectivity, the requirement of fluorescent agents to label the samples, add time and cost to the method which led to the development of a variant of FOBS, tapered fiber optical sensors (TFOS) which confer the advantages of chemical inertness, compatibility to various surface modifications and low cost (82). Another optical sensing technique employed for the bio-receptor-analyte binding detection is label-free surface Plasmon resonance (SPR). SPR sensing system developed by Biacore (GE healthcare) received commercial success in 1990 (83). SPR system allows the plane-polarized light to pass through a glass prism, whose bottom contacts the bio-receptor-immobilized transducer surface made of a thin film of gold. The immobilized bio-receptor is then allowed for the specific binding with the analyte passing through a microfluidic system. This interaction induces the change in the refractive index of the transducer surface which is altering the angle of light exiting the prism (the SPR angle) which is detected further. Various SPR-based biosensors have been developed for the diagnosis of an array of bacterial infections using a range of bio-receptors, such as antibodies (84, 85), lectins (86, 87) and bacteriophages (88, 89). The sensitivity of SPR-based bacterial sensors has been improved by the modification of the transducer surface using nanorods (90). A variant of SPR, Localized surface Plasmon resonance (LSPR) has been developed which employs the use of noble metal nanoparticles to enhance the sensitivity of the system (91). Surface-enhanced Raman scattering (SERS) is yet another modification of optical biosensors that can be used to detect bacterial cells in blood medium even in the slightest concentration. The technique uses roughened metal nano-surfaces (gold or silver) which when excited, creates a highly localized field as the nanostructures drive surface charges in resonance which leads to the enhancement in the Raman signals when a molecule is absorbed or lie close to the field (92). Although the adaption of SPR based systems for point-of-care diagnostics is still limited due to its large size and cost. (Figure 6) Miniaturized SPR-based biosensor (Spreeta SPR chips) by Texas Instruments Inc. has recently been developed but due to the requirement of a microfluidic system, the technique is confined to the laboratory.
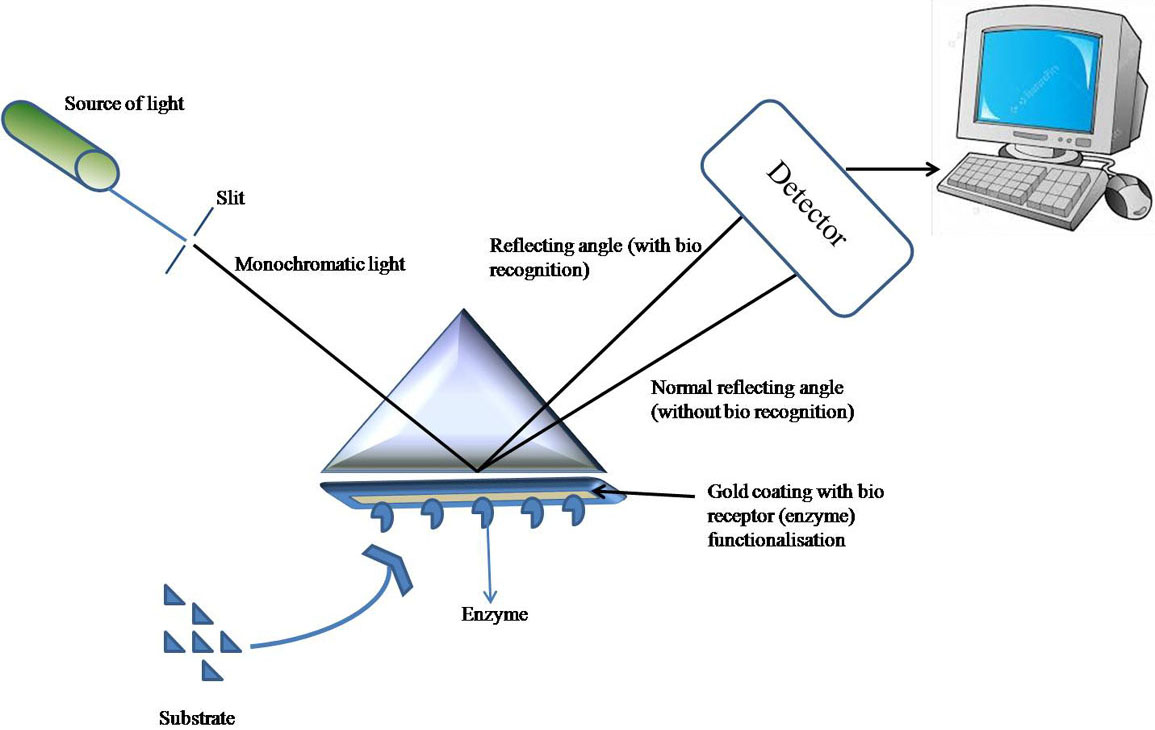 Figure 6
Figure 6Pictorial representation of SPR Biosensor.
Mechanical biosensors work on the principle of addition of a detectable amount of mass which deforms or displace the device and is subsequently detected by converting the applied force into measurable displacement. The mechanical sensors provide high sensitivity (can detect the applied force of up to 10pN), ease of miniaturization, quick analysis time (less than one minute) and no sample processing (93). The mechanical biosensors function centrally on the cantilever which is sensitive to the bound analyte (94). Mechanical biosensors are classified into surface stress and dynamic biosensors depending on whether the device measures quasistatic deflection (surface stress) or oscillation of resonant frequency (dynamic) (95). The widely used mechanical biosensors are QCM sensors which are label-free piezoelectric biosensors. The binding of the analyte to its specific bio-receptors results in the increased mass on the sensor surface which further changes the resonance frequency detected by the sensor. QCM sensors have already been developed for the detection of Vibrio cholerae (96), Escherichia coli (97, 98), Salmonella enterica serovar Typhimurium (99, 100), Francisella tularensis (101), Campylobacter jejuni (102) and Bacillus anthracis (103, 104, 105). The recently developed piezoelectric-excited millimeter-sized cantilever (PEMC) sensors are constructed with the additional piezoelectric layer (made of lead (Pb), zirconate (Zr) and titanium (Ti) referred to PZT) therefore can both excite and sense. This new and improved class of mechanical biosensors has been able to detect E. coli, Bacillus anthracis (106), foodborne pathogens (Listeria monocytogenes) (107) and single-stranded DNA to the femtogram sensitivity (108).
Electrochemical biosensors comprise potentiometric, amperometric, and impedimetric sensing techniques. Amperometric biosensors were the first type of electrochemical biosensors to be described in 1953 (109). Electrochemical biosensors have gained worldwide commercial success, largely due to the development of amperometric glucose sensors in diabetic diagnosis and monitoring (110). The key advantages of the electrochemical biosensors are point-of-care testing, low cost, and miniaturization capacity (111).
Potentiometric biosensors are constructed by combining the specific biological interactions with ion-selective electrodes (transducer) to measure the variation of the potential of a solution. The change in potential is dependent on analyte recognition which is logarithmically proportional to the analyte concentration at the working electrode. The change in potential is detected by measuring the release or absorption of charged anions or cations which is given by the equation FBS-25-4872-g001.jpg where E is the measured potential, Eo is constant for ion-selective electrode, (I) is the concentration of free ions (82). Potentiometric biosensors have been developed for penicillin (H+) (112), glucose (H2O2) (108) and urea (NH4+) (113). Potentiometric stripping analysis (PSA) or chrono-potentiometric method is the technique of determining the metal traces by electrolytic deposition on the electrode and stripping to obtain the stripping potential. The potential stripping is accomplished by using chemical oxidant (Hg (II) or DO) which leads to the oxidation of deposited metal at a constant rate and the rate is determined by the diffusion of oxidant towards the electrode surface. Thus, the signal is recorded as the function of time and the result is deduced in the form of a curve. The distance between two consecutive points in the curve is recorded to be proportional to the concerned metal (114). Marine pathogenic sulfate-reducing bacteria (sulfide detection), Staphylococcus aureus (label-free) have been detected using PSA (115). Although the detection range of PSA is good, the pre-incubation steps are not suitable for rapid and on-site detection requirements. Another technique that has gained importance is the use of a single-walled carbon nanotube (SW-CNT) based aptamer system to measure the real-time electromotive force (EMF) related to bacterial binding to nanotube bound aptamer which generates the concentration-dependent linear signal.
Amperometric biosensors work on the principle of generation of current to measure redox reactions produced in response to analyte-bio-receptor interaction. The interactions used in the bio-recognition are mainly antigen-antibody, enzyme-substrate and DNA-DNA interaction (116). As the concentration of analyte increases, the interaction of analyte-bio-receptor increases and thus the current generated also increases (110). About 4 decades earlier, the enzyme-based amperometric sensor for sensing glucose was introduced which has been applied to a wide range of analytes (117). The advantages of amperometric biosensors are their relative simplicity, sensitivity, ease of miniaturization, and specificity. Therefore, in the field of bio-sensing, amperometric bio-sensing is the most commonly used detection method. The amperometric detection has been done by using wide range of bio-receptors for their specific analytes including antibodies with or without conjugated magnetic nanoparticles, phages and nanocomposites of ferrocene (Fc), fullerene (C60) and thiolated chitosan (118, 119) immobilized on a screen-printed electrode (composed of working electrode, counter electrode, and reference electrode). Amperometric sensors have already been developed for the diagnosis of whole cells (E. coli and Staphylococcus aureus) and intracellular molecules (β-d-galactosidase (120), protein A (121), the specific virulent gene (122).
The term impedance was coined by Oliver Heaviside in the late 19th century and since then, electrochemical impedance spectroscopy (EIS) has been employed for the characterization of biological systems (123). Impedimetric biosensors function by the interaction of an analyte with bio-receptor which in turn causes a change in electron transfer resistance across the working electrode surface. As the concentration of analyte increases, the binding increases proportionately and the impedance change is detected by a transducer (124). The commonly used receptors are antibodies; however, other molecules such as enzymes and nucleic acids may also be used which are capable of detecting a wide range of analytes (125,126). Impedance (Z) is a function of two components viz. Resistance (R) and capacitance (C) can be represented as Nyquist plots where R (a Real component of impedance) is plotted on the x-axis and C (an imaginary component of impedance) is plotted on the y-axis. The Nyquist plot can be interpreted as a semicircular plot with an observation of 45o rise where the major component of impedance is derived from the solution resistance (Rs) at high frequency however at lower frequency, impedance arises from the charge transfer resistance (Rct) described as resistance to the flow of electrons to the electrode surface. Commonly, the data is presented by recording the change in Rct upon analyte addition; however, real impedance, imaginary impedance, or absolute impedance against bacterial concentration may also be plotted. Chrono-impedimetric data can also be obtained by taking measurements at a fixed frequency to monitor real-time binding (127-129). The increasing deposition on the sensor surface leads to the rise in the change in impedance, either layer-by-layer sensor construction or analyte binding and the plot is plotted quantitatively. Impedimetric detection of an analyte can be classified into faradic and non-faradic impedance based on presence or absence of the use of an additional redox mediator (Ru(NH3)63+/2+ (hexa-ammineruthenium III/II ions) and Fe(CN6)3−/4− (ferricyanide/ferrocyanide)). The complexity of impedance measurement depends on the choice of electrode material, base layer construction (a type of self-assembled monolayer SAM or polymers), bio-receptor conjugation chemistry, choice of analytes (type and size), and complexity of the sample matrix. These issues have turned the research focus toward optimizing layer-by-layer sensor construction to achieve the optimum impedance signal with minimum noise. The sensor construction varies widely in the selection of base electrode materials, choice of bio-receptor, linking chemistry, and finally impedance data representation. These biosensors are a very promising choice for the microbial diagnosis as they are label-free, inexpensive, highly sensitive, specific, easy to miniaturize and allows unrestricted measurement of the molecule of interest i.e., no requirements for the formation of electroactive species). A plethora of reports discusses the impedimetric detection of E. coli, Salmonella typhimurium, Campylobacter jejuni, and Staphylococcus aureus. Currently, no impedance biosensors had widespread success commercially, although the use of this technology is increasing rapidly. Apart from having the number of advantages of impedance biosensors, variable reproducibility, the high limit of detection, and problems with nonspecific binding are cited as disadvantages of impedance biosensors (124,125). Despite having disadvantages, the technology is constantly improved, thus, EIS becomes an increasingly attractive technique in biosensor applications. (Figure 7)
 Figure 7
Figure 7Schematic representation of electrochemical biosensor based on Nucleic acid probes.
A label-based lateral flow dipstick assay for the rapid and simple detection of multiplex loop-mediated isothermal amplification (m-LAMP) was developed for leptospira diagnosis. The assay was developed for the simultaneous detection of the target DNA template and a LAMP control. This biosensor for the detection of pathogenic leptospira operates through a system where probe-hybridization and the additional incubation steps are eliminated. The lateral flow dipstick was developed to detect three targets, the LAMP target amplicon, and the LAMP internal control amplicon and chromatography control. The result of the appearance of three lines on the dipstick, indicated positive results for all representative pathogenic Leptospira species, whereas two lines indicated negative results, for other bacterial species. The specificity of this biosensor assay was found to be 100% when it was tested with 13 representative pathogenic Leptospira species, 2 intermediate Leptospira species, 1 non-pathogenic Leptospira species, and 28 other bacterial species. This particular DNA biosensor was able to detect DNA at concentrations as low as 3.95 × 10−1 genomic equivalent ml−1. An integrated m-LAMP and label-based lateral flow dipstick were successfully developed, promising simple and rapid visual detection in clinical diagnostics and serving as a point-of-care device (130). Furthermore, an electrochemical DNA sensor has been developed for leptospirosis which is highly sensitive and specific towards the target organism. The sensor was developed by targeting lipL32 gene which is highly conserved among pathogenic serovars of Leptospira. The biosensor is based on AuNPs modified multiwalled carbon nano tubes immobilized with amino labeled ssDNA probe specific to lipL32 gene. The biosensor was reported with the sensitivity of 264.5 μA/cm2/ng, lower limit of detection (LOD) of 0.015 ng/6μl with stability of 6 months at 4ºC with only 13% loss in Ip value (131).
Although a variety of conventional techniques have been developed to diagnose leptospirosis but sensitivity, reproducibility, and ease of miniaturization before their successful translation forms key challenges to be addressed. Because the national and international severity and the mortality rate of the disease is alarming in different coastal regions, so there should be some accurate and specific point of care systems. Bio-sensing shows great promise in the field of disease diagnosis, being highly sensitive and label-free. However, research needs to be conducted with an emphasis on reproducible, inexpensive, and novel electrode material, stable conjugation, and strict optimization of bio-receptor configuration, orientation, and concentration. Miniaturization of biosensing systems and robotic layer-by-layer construction will ultimately improve sensor performance with high reproducibility for commercialization. As glimpsed from the article, there are infinite studies of the detection of harmful microbes from food, environment, and infections. Leptospira is one of the organisms that require an immediate diagnostic assay as the existing gold standards for the disease is proven imperfect and bio-sensing paves a novel way to diagnose the organism and allow the suspected patients to receive the treatment as soon as possible so that they could be saved from losing their lives to this deadly organism. So amperometric biosensor is showing a great impact in the field of diagnostics due to its higher sensitivity and can become a very useful tool in the diagnosis of leptospirosis. Aptamer based sensors can also become suitable alternative for the diagnosis due to its higher affinity towards target. So this review summarizes about the prevalence, pathophysiology and latest techniques including biosensors that can be used in accurate diagnosis of leptospirosis.
The author acknowledges the Indian Council of Medical Research, Govt. of India for their grant- Leptos/27/2013-ECD-1.
WHO
world health organization
global burden of disease
cerebrospinal fluid
polymerase chain reaction
enzyme linked immunosorbent assay
microscopic agglutination test
sexually transmitted infection
surface Plasmon resonance
electrochemical impedance spectroscopy
