Thyroid hormone (T3) is important for adult organ function and vertebrate development. Amphibian metamorphosis is totally dependent on T3 and can be easily manipulated, thus offering a unique opportunity for studying how T3 controls vertebrate development. T3 controls frog metamorphosis through T3 receptor (TR)-mediated regulation of T3 response genes. To identify direct T3 response genes, we previously carried out a ChIP (chromatin immunoprecipitation)-on-chip analysis with a polyclonal anti-TR antibody on the tadpole intestine and identified many putative TR target genes. Among them is the methyl-CpG binding domain protein 3 (MBD3) gene, which has been implicated to play a role in epigenetic regulation of cellular processes as a subunit of the Mi-2/NuRD (Nucleosome Remodeling Deacetylase) complex. We show here that MBD3 is upregulated in the intestine and tail by T3 and its expression peaks at stage 62, the climax of metamorphosis. We further show that a putative TRE within the first intron of the MBD3 gene binds to TR/RXR in vitro and in vivo, and mediates T3 regulation of the MBD3 promoter in vivo.
Thyroid hormone (T3) is important for proper development and normal physiology of many adult organs/tissues in vertebrates (1; 2; 3; 4; 5). T3 deficiency during human development results in significant pathological consequences such as the formation of human cretins, who are short in stature and severely mentally retarded (6). The most critical period of T3 action in mammals is the postembryonic period, which covers several months around birth when T3 levels are high in the plasma (1). Owing to difficulty to access and/or manipulate the uterus-enclosed late stage of mammalian embryos and neonates, it has been difficult to study how T3 affect mammalian postembryonic development.
Frog metamorphosis mimics mammalian postembryonic development (1; 2; 7). Similar to mammalian postembryonic development, frog metamorphosis involves distinct changes in different organs and tissues. During frog metamorphosis, larval specific organs, such as the tails and gills, are totally resorbed while adult specific tissues such as the limbs develop de novo. The majority of organs/tissues are present in both larval and adult stages but undergo extensive remodeling during metamorphosis. A well-studied such tissue is the intestine, which undergoes drastic changes involving apoptotic degeneration of vast majority of the larval epithelial cells and concurrent de novo formation of adult stem cells that rapidly proliferate and eventually give rise to a multiply folded adult epithelium resembling mature mammalian intestine (8; 9; 10).
Strikingly, T3 plays a causative role for anuran metamorphosis (1; 2; 7). T3 exerts its effects by regulating target gene expression through T3 receptors (TRs). For genes induced by T3, TRs can form heterodimer with 9-cis retinoic acid receptors (RXRs) and bind to T3-response promoters to repress or activate T3-inducible genes depending on the availability of T3 (5; 11; 12; 13; 14; 15; 16; 17). In premetamorphic tadpoles, T3 concentrations are low and the unliganded TRs repress T3-inducible genes by recruiting histone deacetylase-containing corepressor complexes (17; 18; 19; 20; 21; 22; 23; 24; 25; 26; 27). This helps to ensure proper premetamorphic growth and prevent premature metamorphosis. When T3 becomes available during metamorphosis, the liganded TRs recruit histone modifying coactivator complexes to the T3 response genes, leading to histone modifications, chromatin remodeling and gene activation, to effect the drastic tissue-specific metamorphic changes. Recent studies have shown that TR appears to be necessary and sufficient to mediate the metamorphic effects of T3 (17-27).
While much has been learnt about the molecular mechanism of TR action in vitro and in vivo, it is important to determine the downstream events that are responsible for the cellular and morphological changes. Arguably, the most important issue is the identification and functional characterization of direct T3 target genes during metamorphosis. Toward this end, we have been focusing on intestinal metamorphosis, a process that involves both apoptotic degeneration of the larval epithelium and concurrent de novo formation of adult stem cells (8; 9; 10). We have previously carried out a ChIP (chromatin immunoprecipitation)-on-Chip analysis of TR binding in the intestine from premetamorphic tadpoles treated with or without T3 (28). Among many thus identified putative TR target genes is the methyl-CpG binding domain protein 3 (MBD3) gene. MBD3 was originally identified as a protein bearing a highly similar domain to the methyl-CpG binding protein 2 (MeCP2) and a subunit of the Mi-2/NuRD (Nucleosome Remodeling Deacetylase) complex that has both chromatin remodeling and histone deacetylase activities (29; 30; 31). Unlike most Methyl-CpG-binding domain proteins such as MeCP2, MBD1, MBD2 and MBD4 that can bind to methylated DNA, mammalian MBD3 does not bind methylated DNA but binds to hydroxymethylated DNA instead (29; 32; 33), though a Xenopus MBD3-like protein has been shown to bind methylated DNA in vitro (34). Thus, T3 likely induces the expression of MBD3, which in turn affects downstream events via an epigenetic pathway involving the Mi-2/NuRD complex.
To determine whether and how MBD3 is regulated by TR directly at the transcriptional level, we have carried out a bioinformatics analysis of the region bound by TR from the ChIP-on-chip assay and identified two putative TREs. We show that MBD3 is indeed upregulated by T3 in the intestine during natural and T3-induced metamorphosis. In addition, T3 also regulates its expression in the tail. More importantly, we provide in vivo and in vitro evidence to show that a TRE in the first intron of the MBD3 gene mediates the T3 induction of the gene during metamorphosis.
Xenopus tropicalis and Xenopus laevis were purchased from Nasco (Fort Atkinson, MI). Tadpoles were staged according to Nieuwkoop and Faber (35). When indicated, premetamorphic tadpoles at stage 54 were treated with 10 nM T3 at 25 oC for 2 days. All animal procedures were done as approved by NICHD Animal Use and Care Committee.
Total RNA was isolated from Xenopus tropicalis tadpole intestine and tail at indicated stages during natural metamorphosis or premetamorphic tadpoles at stage 54 treated with or without T3 for 2 days. cDNA was synthesized from 2.0 μg total RNA by using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) in 40 μl reactions according to the manufacturer’s instructions. qRT-PCR was carried out by using SYBR Green PCR Master Mix on a StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA). The primers used were 5’-TCAAGCAACCAGTGACCAAG -3’ (forward) and 5’-TTTCCCAGAAGAGCTGCCT -3’ (reverse) for MBD3. EF1α (elongation factor 1α) was used as the normalization control as described previously (36). Each RNA preparation was from tissues pooled from at least 3 tadpoles and the qRT-PCR was done on duplicated sets of samples.
The sequence of Xenopus tropicalis MBD3 gene was downloaded from ENSEMBL website (https://useast.ensembl.org/index.html) and the computational analysis tool NHR-Scan (http://www.cisreg.ca/cgi-bin/NHR-scan/nhr_scan.cgi) (37) was used to identify the putative TREs.
Gel mobility shift assay was performed essentially as described previously (36; 38). In brief, Xenopus tropicalis TRα and RXRβ proteins were made by using TnT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI). They were mixed with double-stranded, infrared dye IR700 (LI-COR, Lincoln, NE)-labelled TRE oligonucleotide of Xenopus laevis TRβA gene in the in vitro binding reaction in the presence or absence of unlabeled competitors containing the wild type or mutant putative TREs of Xenopus tropicalis MBD3 gene. The unlabeled competitors were made by annealing synthetic, complementary oligonucleotides. The oligonucleotide sequences of the upper strands were 5’-ATTTGTGGTCAGACCAATTCATCCAT-3' (MBD3 TRE1), 5’-ATTTGGGGTCAGATGGGGACATCCAT-3' (MBD3 TRE2), and 5’-ATTTGGAGTCAGATGGAAACATCCAT-3' (MBD3 mTRE2) (bold letters indicate the TRE half sites with the mutated nucleotides underlined in the mutant TRE). Each 20 μl binding reaction included 1 μl of 100 fmol of IR-700 labelled probe, 1 μl each of TR and RXR in vitro translated protein mixture, and 1 μl of the wild type or mutant TRE oligonucleotides at 400 fmol/μl, 2 pmol/μl, or 10 pmol/μl, respectively, to obtain 4x, 20x, or 100x unlabeled competitor oligonucleotides, respectively. The mixtures were incubated at room temperature for 20 min and electrophoresed on a 6% DNA retardation gel (Invitrogen, Carlesad, CA). The resulting gel was then scanned by using an Odyssey Infrared Scanner (LI-COR, Lincoln, NE). The assay was done three times with similar results.
The fragment encompassing 1 kb upstream of the 5’-end of the reported Xenopus tropicalis MBD3 cDNA, the exon 1, and part of intron 1 that contained predicted TRE2, was PCR-amplified from genomic DNA with the primer pair of 5’-cctgagctcGCTAGCGGTGATATCACTCCAACTTGCAGC-3’ (Forward, bearing a NheI site at the 5’-end) and 5’-cggattgccAAGCTTgCTCTTTATTCCTCC-AGCTGCACC-3’ (Reverse, bearing a HindIII site at the 5’-end) by using high fidelity PrimeStar DNA Polymerase (Takara, Mountain View, CA). The PCR fragment was double-digested with NheI and HindIII, gel-purified, and cloned into pre-digested pGL4.10 firefly luciferase vector (Promega, Madison, WI) bearing the same restriction ends. The mutant promoter harboring a mutated TRE was PCR-amplified from the wild type promoter construct DNA by using the same forward primer for the wild type promoter fragment and 5’-tgccaagcttgCTCTTTATTCCTCCAGCTGCACCCAGTCTGTATGTTTCCATCTGACTCAC-3' (bearing a HindIII site at the 5’-end with the mutated nucleotides underlined). The PCR fragment was double-digested with NheI and HindIII, gel-purified and cloned into pre-digested pGL4.10 vector bearing the same restriction ends. The mutant construct was confirmed by DNA sequencing.
Oocyte transcription assay was performed as described (38; 39). Briefly, the plasmid constructs containing the coding region of GFP, Xenopus tropicalis TRα and RXRβ were linearized with EcoRI digestion and transcribed by using a mMESSAGE mMACHINE T7 Transcription Kit (Ambion, Grand Island, NY), respectively. The cytoplasm of stage VI Xenopus laevis oocytes was injected with 46 pg/oocyte of GFP mRNA or TR/RXR mRNA mixture. Two hour later, the reporter construct in which firefly luciferase was under the control of MBD3 promoter harboring the wild type or mutated TRE was injected into the nuclei of these oocytes (33 pg/μl) along with the internal control Renilla luciferase reporter phRG-TK (3.3 pg/μl). After incubation at 18 oC overnight in the presence or absence of 100 nM T3, groups of oocytes were collected for dual luciferase assay following the manufacturer’s instructions for the Dual-Luciferase-Reporter Assay kit (Premega, Madison, WI). The relative expression of the firefly luciferase to Renilla luciferase was determined and presented as the average of at least three groups of oocytes. The data shown were representative of a few independent experiments with similar results.
ChIP assay on Xenopus tropicalis tadpole intestines was done as described previously (40) with an antibody against TR (anti-TR) (38), RNA Polymerase II (abcam, Cambridge, MA), or methylated histone H3K79 (anti-H3K79m2, abcam, Cambridge, MA), and with IgG as a negative control, by using Chromatin Immunoprecipitation (ChIP) Assay kit (Millipore, Burlington, MA). Each sample had three replicas and each replica included at least 5 tadpoles. The immunoprecipitated DNA was analyzed by qPCR with SYBR Green PCR Master Mix on a StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA). For the analysis of MBD3 TRE, primers 5’-ATGCCCGCCTACTCTTTATTCCTCCAGCTGC-3’ (forward) and 5’-GAGAGAGAGTCAGTGTGGTGGTGGGTCAGA-3’ (reverse) were used. All ChIP experiments were done twice with similar results.
Our ChIP-on-Chip assay identified TR association with MBD3 gene in the tadpole intestine (28). To investigate whether MBD3 is regulated by T3 in tadpoles, we treated premetamorphic Xenopus tropicalis tadpoles at stage 54 with or without 10 nM T3 for 2 days and analyzed MBD3 gene expression by qRT-PCR on total RNA isolated from the intestine and tail, which undergoes resorption instead of remodeling during metamorphosis. The data showed upregulation of MBD3 expression by T3 in both organs, although less dramatically in the tail (Figure 1), suggesting that MBD3 is indeed a T3 target gene and that its expression should also increase during natural metamorphosis when T3 level is high. Indeed, when we analyzed its expression by qRT-PCR in the intestine and tail at different stages from premetamorphic (stage 56), metamorphic climax (stage 58-64), to the end of metamorphosis (stage 66), we observed that MBD3 expression was significantly upregulated in the intestine during natural metamorphosis with a peak expression at stage 62 (Figure 2A) when T3 level in the plasma is high and the intestine undergoes drastic remodeling, including rapid larval intestinal epithelial degeneration through apoptosis and robust adult intestinal stem cell proliferation and differentiation (8; 9; 10). In addition, MBD3 expression in the tail also increased dramatically when the tail underwent rapid resorption around stage 62 to 64 (Figure 2B). Thus, MBD3 is upregulated by T3 and likely participates in the cell fate determination during intestinal remodeling and tail resorption during frog metamorphosis.
 Figure 1
Figure 1
MBD3 expression is up-regulated by T3 treatment of premetamorphic Xenopus tropicalis tadpoles. Intestinal (A) and tail (B) RNAs were extracted from premetamorphic tadpoles at stage 54 treated with or without 10 nM T3 for 2 days and subjected to cDNA synthesis. qRT-PCR was conducted to examine the expression of MBD3 and control gene EF1α (elongation factor 1α) mRNAs, respectively. MBD3 expression was normalized against that of EF1α in the same cDNA samples and presented as mean +/- standard error of the mean (S.E.M). Unpaired t-test with Welch’s correction was done to compared the two samples (* p<0.05).
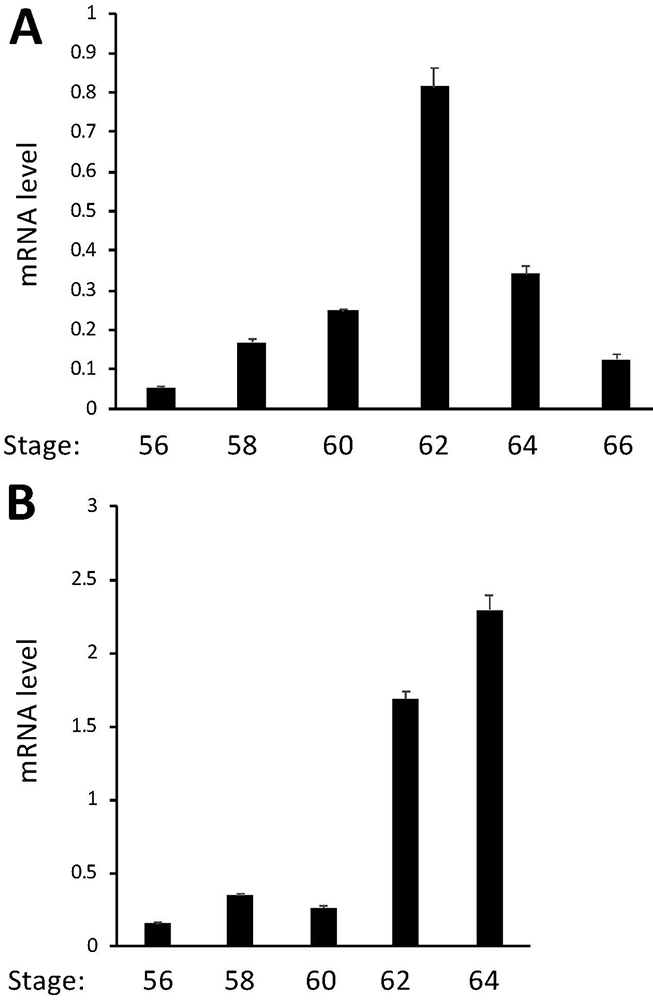 Figure 2
Figure 2
MBD3 expression peaks at the climax of metamorphosis in the intestine and tail during natural metamorphosis. Intestinal (A) and tail (B) RNAs were extracted from metamorphosing tadpoles and subjected to cDNA synthesis. qRT-PCR was conducted to examine the expression of MBD3 and control gene EF1α, respectively. MBD3 expression was normalized against that of EF1α in the same cDNA samples and presented as mean +/- standard error of the mean (S.E.M). One-way ANOVA followed by Boferroni’s test was done to compare the samples (* p<0.05).
As our ChIP-on-Chip analysis showed that TR was associated with MBD3 gene in the tadpole intestine, we carried out bioinformatics analysis on the sequences around the transcriptional start site, which include the region associated with TR found from the ChIP-on-Chip analysis, by using NHR-Scan and identified two putative TREs. One TRE is upstream of the predicted transcription start site (TRE1) and the other one in the first intron (TRE2, Figure 3A), with the TRE2 having a sequence more conserved with the consensus TRE made of two direct repeats of AGGTCA half site separated by 4 bp (Figure 3B). To investigate if TR/RXR heterodimer binds to these two putative TREs, we carried out in vitro gel mobility shift assay by using as the probe of IR700-labelled TRE of Xenopus laevis TRβA gene, a well characterized TRE consisting of two near perfect direct repeats of AGGTCA half site separated by 4 bp (Figure 3B) (39; 41), TR and RXR proteins made through in vitro translation, and unlabeled competitor TREs made from sequences of the putative Xenopus tropicalis MBD3 TREs (Figure 3B). The results indicated that the intronic TRE (TRE2) of the MBD3, but not the TRE1 in the upstream region, competed for binding to TR/RXR heterodimer strongly (Figure 3C). To determine if the strong binding of TRE2 to the TR/RXR heterodimer was sequence-specific, we mutated the putative TRE2 at the positions known to be important for binding to TR/RXR heterodimer (Figure 3B) and determined the effect in the mobility shift assay. The results shown in Figure 3C indicated that the mutations in the TRE2 abolished its ability to compete for binding to TR/RXR heterodimer. Thus, the intronic TRE2 of Xenopus tropicalis MBD3 binds to TR/RXR heterodimer specifically and likely mediates liganded TR activation of MBD3 gene expression during frog metamorphosis. Given the inability of the TRE1 to bind to TR/RXR in vitro, we focused the remaining studies on TRE2.
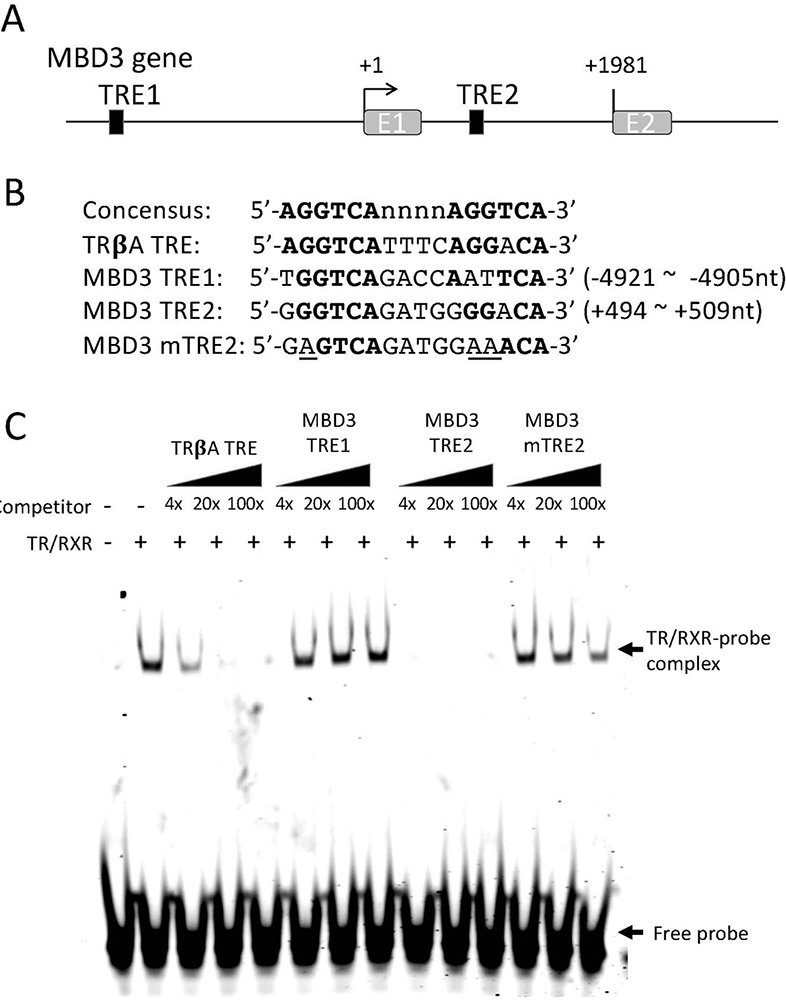 Figure 3
Figure 3
TR binds to a putative TRE within the first intron of the MBD3 gene in vitro. (A) Schematic diagram showing two putative TREs (solid boxes) in X. tropicalis MBD3 gene. (B) The sequences of the putative TREs are shown in comparison with the sequences of consensus TRE and the TRE of Xenopus laevis TRβA gene (TRβA TRE). The numbers indicate the locations relative to the transcriptional start site of MBD3 gene based on the 5’-terminus of the reported MBD3 cDNA sequence (denoted as +1). A mutant version of MBD3 TRE2 (MBD3 mTRE2, with the mutated nucleotides underlined) was made for TR/RXR binding assay in comparison with the wild type TREs. Bold letters indicate nucleotides conserved with the consensus TRE. Note that the putative TRE1 (MBD3 TRE1) is in the 5’-proximity of its promoter and the putative TRE2 (MBD3 TRE2) is in the first intron of the gene. E1: exon1; E2: exon2. (C) The putative MBD3 TRE2 but not TRE1 compete for binding to TR/RXR heterodimers against labeled TRβA TRE in a gel mobility shift assay. The labelled double stranded probe containing TRβA TRE was mixed with in vitro translated TR/RXR heterodimers in the presence or absences of 4x, 20x, or 100x of unlabelled wild type or mutant MBD3 TREs as indicated. The reaction mixtures were analyzed by electrophoretic mobility shift assay (EMSA). The mock in vitro translation mixture was used as a negative control for TR/RXR heterodimers (lane 1 on the left). The locations of TR/RXR bound probe (TR/RXR-probe complex) and unbound probe (Free probe) are indicated with arrows. Note that MBD3 TRE1 had little competition against the labelled TRβA TRE probe while MBD3 TRE2 strongly competed against the labelled probe. Mutations in the MBD3 TRE2 drastically reduced its ability to compete against the labeled probe for TR/RXR binding.
To determine if the intronic TRE2 can mediate the transcriptional regulation of Xenopus tropicalis MBD3 expression by T3 in vivo, we cloned a genomic MBD3 fragment encompassing 1 kb upstream of the predicted transcription start site, exon 1 (276bp) and a part of intron 1 that included the putative TRE2 (265bp), upstream of the coding region of firefly luciferase gene in a reporter construct (Figure 4A). The transcription from the MBD3 promoter construct would produce an mRNA consisting of MBD3 exon 1 (152 bp for 5’-untanslated region, 108 bp for encoding a N-terminal fragment of the MBD3 protein), 265 bp of intron 1, and the coding sequence for the firefly luciferase, which encodes a fusion firefly luciferase protein containing 137 additional amino acids at its N-terminus. As a control, we also generated a mutant construct where TRE2 was mutated as in Figure 3B.
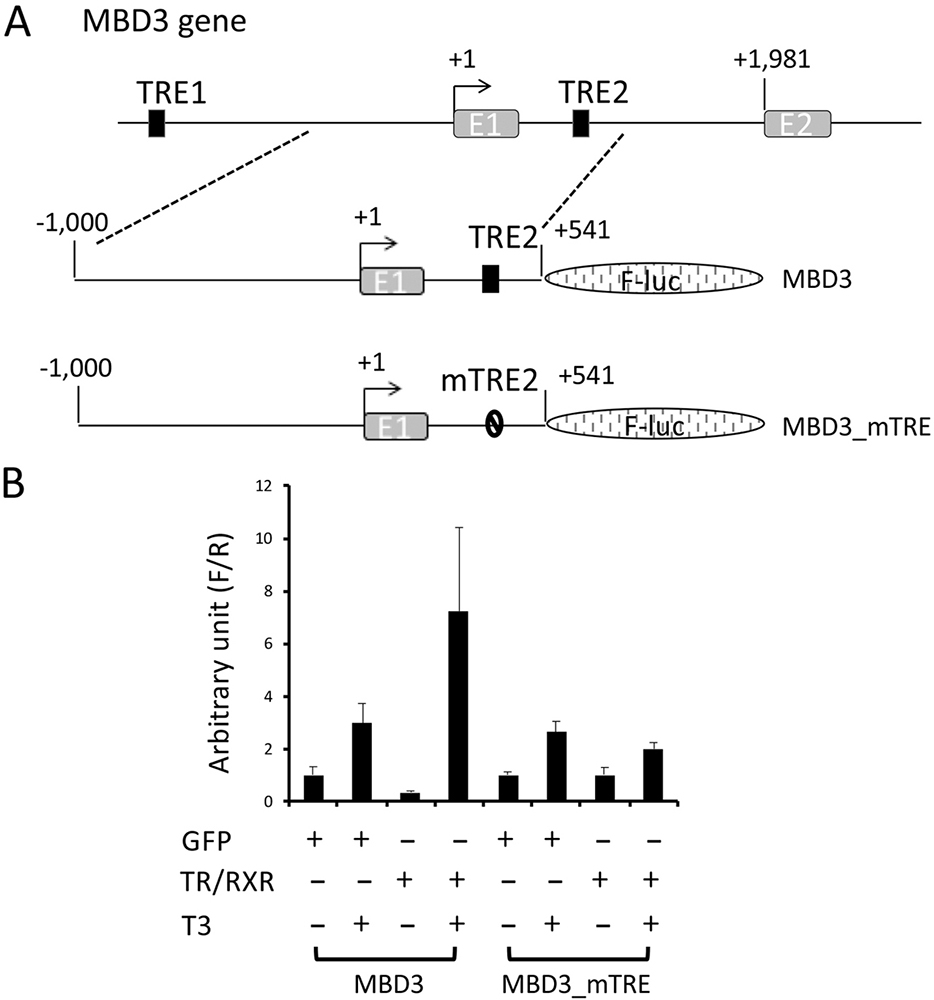 Figure 4
Figure 4
Activation of Xenopus tropicalis MBD3 promoter by liganded TR in vivo is dependent on the TRE2 within the first intron. (A) Schematic diagram of the MBD3 promoter construct with the putative intronic TRE2 (MBD3) or a mutant TRE2 (MBD3_mTRE). The wild type and mutant promoter fragments were cloned in front of the firefly luciferase coding region. (B) The MBD3 promoter is activated by liganded TR in frog oocytes. The wild type or mutant promoter construct was co-injected with the control Renilla luciferase construct phRG-TK into the nuclei of Xenopus laevis oocytes with prior cytoplasmic injection of GFP mRNA or Xenopus tropicalis TRα and RXRβ mRNAs. The oocytes were incubated at 18 oC overnight in the absence or presence of 100 nM T3 and then lysed for dual luciferase assay. The relative activities of the firefly luciferase to Renilla luciferase were plotted. Error bars indicate S.E.M. Note that mutations in the intronic TRE abolished the liganded TR-mediated activation of the promoter. One-way ANOVA followed by Boferroni’s test was done to compare the samples (* p<0.05, ns: not significant).
To study the promoter activity of the reporter constructs, we employed a reconstituted Xenopus laevis oocyte transcription system, which allows the analysis of the promoter activity in the context of chromatin in vivo (39). First, we microinjected mRNAs encoding GFP or TR and RXR into the cytoplasm of Xenopus laevis oocytes to allow the synthesis of the proteins. Then, we microinjected the MBD3 firefly luciferase reporter construct and an internal control plasmid phRG-TK driving the expression of Renilla luciferase into the nuclei of the oocytes. After overnight incubation of the oocytes in the presence or absence of T3, they were harvested for dual luciferase activity assays. The ratio of firefly luciferase to Renilla luciferase activities was then determined as the activity of the MBD3 promoter. As shown in Figure 4B, T3 treatment led to a small increase in the activity of the wild type MBD3 promoter in the absence of overexpressed TR/RXR. When TR/RXR mRNAs were injected into the oocytes, the reporter activity was reduced slightly in the absence of T3, but was strongly activated when T3 was present (Figure 4B). When the intronic TRE2 was mutated, the resulting mutant MBD3 promoter was not affected by the presence of TR/RXR (Figure 4B). Thus, the intronic TRE2 can mediate transcription regulation by liganded TR/RXR in vivo.
Our ChIP-on-Chip assay suggested that TR was bound to the proximity of MBD3 promoter region in the intestine of premetamorphic Xenopus tropicalis. To determine if TR is bound to the intronic TRE2 in vivo, we carried out ChIP assay on intestinal chromatin extracted from premetamorphic tadpoles at stage 54 treated with or without T3 for 2 days by using a polyclonal anti-TR antibody (anti-TR). Analysis of the ChIP DNA by using a primer pair flanking TRE2 showed that TR was bound to TRE2 in premetamorphic tadpole intestine and T3 treatment enhanced TR binding to TRE2 (Figure 5).
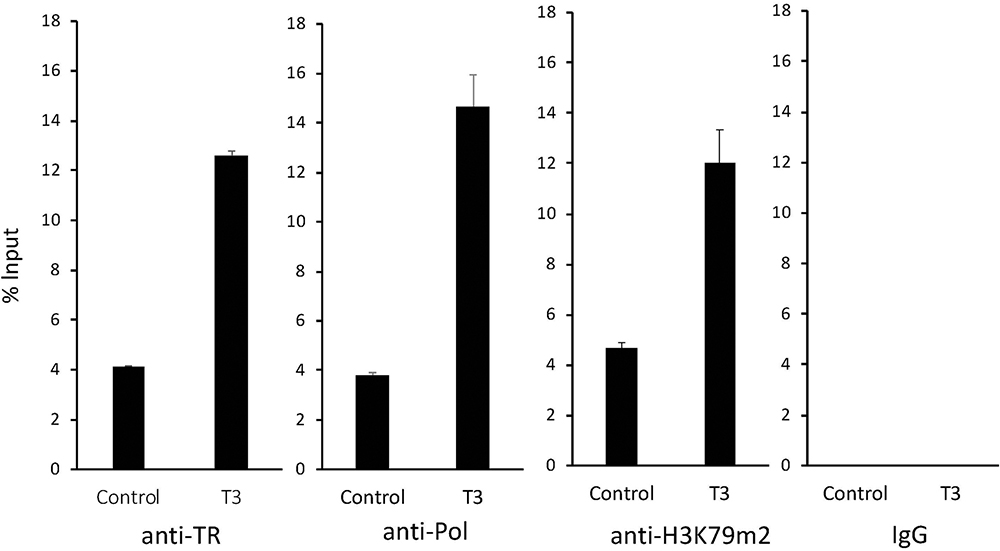 Figure 5
Figure 5
TR binds to the intronic TRE of MBD3 gene in the intestine during T3-induced metamorphosis. Premetamorphic tadpoles at stage 54 were treated with (T3) or without T3 (Control) for 2 days. The intestines were dissected out for ChIP assays with a polyclonal antibody against frog TR (anti-TR), RNA polymerase II (anti-Pol), and histone H3 K79m2 (anti-H3K79m2), or negative control IgG (IgG). The immunoprecipitated DNA was analyzed by qPCR for the enrichment of the intronic TRE2 region of the MBD3 gene (MBD3 TRE). Note that strong binding of TR to the MBD3 intronic TRE was observed in the absence of T3 in the premetamorphic tadpoles, and this binding was increased significantly upon T3 treatment. T3 treatment also drastically enhanced the recruit of RNA polymerase II and the level of H3K79 methylation.
To investigate if this TR binding was important for the activation of the MBD3 gene, we carried out ChIP assay with an antibody against RNA Polymerase II to measure transcription of the gene, and an antibody against methylated histone H3K79 (H3K79m2), a well-known activation histone mark associated with gene activation by TR during metamorphosis (40; 42). The results showed that T3 treatment led to strong increase in both RNA polymerase II recruitment and H3K79 methylation around TRE2 (Figure 5). In contrast, when ChIP assay was done with the negative control IgG, no signal was detected around the TRE2 region (Fig.5). Thus, the intronic TRE2 in the MBD3 gene is bound by TR in premetamorphic tadpole intestine and likely mediate the activation of MBD3 expression by T3 via histone modification during metamorphosis.
Amphibian metamorphosis offers an excellent opportunity to study the molecular pathways underlying postembryonic vertebrate development largely because of the easiness with which to access and manipulate metamorphosing tadpoles. It has been shown that TR plays an essential and sufficient role in mediating the causative effects of T3 during amphibian metamorphosis (18; 27). TR functions by recruiting cofactor complexes to activate and repress T3-inducible genes in the presence and absence of T3, respectively. Thus, it becomes critical to identify and functionally characterize T3 response genes during metamorphosis. Our earlier ChIP-on-Chip analysis has led to the discovery of MBD3 as a putative direct target gene of TR during intestinal metamorphosis. Here we have shown that MBD3 is indeed regulated by T3 during metamorphosis with its expression upregulated not only in the intestine but also in the tail during T3-induced or natural metamorphosis. More importantly, we have provided evidence to support a TRE located in the first intron of the MBD3 gene that binds to TR to facilitate the local histone modification and RNA polymerase II recruitment to activate MBD3 transcription in the presence of T3.
Earlier ChIP-on-Chip analysis showed that TR was associated with MBD3 gene in the intestine of premetamorphic Xenopus tropicalis tadpoles. Our bioinformatics analysis discovered two putative TREs flanking the predicted transcription start site. The putative TRE1 found in the upstream of MBD3 transcriptional start site had no detectable binding to TR/RXR heterodimer in vitro, suggesting it unlikely a functional TRE in vivo. In contrast, the TRE located in the first intron, TRE2, can bind to TR/RXR heterodimers in vitro and is required for the activation of the MBD3 promoter in the reconstituted Xenopus oocyte transcription system in vivo. More importantly, TRE2 is bound by TR in the tadpole intestine and T3 treatment of premetamorphic tadpoles leads to increased level of the activation histone mark H3K79 methylation and recruitment of RNA polymerase II to the TRE2 in the tadpole intestine. Thus, TRE2 is likely responsible for mediating direct transcriptional activation of the MBD3 gene by liganded TR/RXR.
Consistent with being a direct target gene of TR, MBD3 mRNA level is upregulated in the intestine during natural metamorphosis, with peak levels occurring at the climax of metamorphosis when T3 levels are high. In addition, its mRNA level also increases dramatically during tail resorption. Furthermore, T3 treatment of premetamorphic tadpoles leads in the upregulation of MBD3 gene in both the tail and intestine. Thus, it is very likely that MBD3 is important for diverse transformations in different tissues/organs during metamorphosis, including larval cell death and adult cell development and proliferation.
MBD3 contains a domain highly similar to the methyl-CpG binding protein 2 (MeCP2) and is an essential scaffold subunit of the Mi-2/NuRD complex that can deacetylate histones and remodel chromatin upon ATP hydrolysis (29; 30; 31). The Mi-2/NuRD complex is a unique player among chromatin remodeling complexes because it couples ATP-dependent nucleosome remodeling activity with histone deacetylase activity (30; 32; 43; 44; 45). Mammalian MBD3 does not bind methylated DNA but binds to hydroxymethylated DNA instead (29; 32; 33), although a Xenopus laevis MBD3-like protein can bind methylated DNA in vitro (34). Thus, it is likely that MBD3 may affect amphibian metamorphosis through Mi-2/NuRD complex via binding to methylated or hydroxymethylated DNA.
MBD3 has been shown to affect development and stem cells in mammals. Depleting MBD3 coupled with OSKM (Oct4, Sox2, Klf4 and Myc) transduction and reprogramming result in deterministic and synchronized iPS cell programming from mouse and human cells (46), though other data have also shown that a profound reduction in reprogramming efficiency was observed from cells where MBD3 had been ablated (47). Nevertheless, MBD3 likely can affect induction of cell pluripotency and stem cell lineage commitment. In addition, regulating MBD3’s stability and MBD3/NuRD complex recruitment has been implicated to affect neural progenitor cells’ self-renewal and neuronal differentiation during mammalian corticogenesis (48). MBD3/NuRD has also been reported to be required to repress inappropriate transcription in both progenitor cells and neurons to facilitate appropriate cell lineage choice and differentiation programs during mouse neurogenesis (49). MBD3 is critical for mouse development with a distinct role from even its closely related protein MBD2. MBD3 knockout (MBD3-/-) mice die during early embryogenesis whereas MBD2 knockout (MBD2-/-) mice are viable and fertile (50). MBD3-/- embryonic stem cells misregulate a subset of pluripotency-associated genes and subsequently fail to engage in cell differentiation into embryonic lineage when self-renewal requisites are withdrawn from the culture media (43; 51; 52). The upregulation of MBD3 during both intestinal remodeling, where larval cell death and adult stem cell development take place, and tail resorption, where all tissues degenerate, would argue for a critical role of MBD3 in larval cell death as well as adult organogenesis during metamorphosis, consistent with the essential role of MBD3 during mouse development. Clearly, it would be interesting to determine if MBD3 is expressed in a cell type/tissue-specific manner during metamorphosis and what roles the endogenous MBD3 plays by taking advantage of the ability to carry out gene knockout in Xenopus tropicalis and laevis (53; 54- 56).
This work was supported by the intramural Research Program of NICHD, NIH. LF and CL conceived and designed the experiment; WN provided materials; LF and YS analyzed the data and prepared the manuscript; all authors approve the final version of the manuscript. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
