Academic Editor: Yiannis Kourkoutas
Background: The vineyard is a great reservoir of autochthonous yeast strains whose composition is defined by different regional (edaphology, orography or climatology) and anthropological factors (cultivation systems or cultural practices). Most of this yeast diversity corresponds to non-Saccharomyces strains, some of which have potential use in winemaking. Methods: The oenological potential of 29 different native non-Saccharomyces strains belonging to 4 species (Lachancea thermotolerans, Torulaspora delbrueckii, Starmerella bacillaris and Metschnikowia spp.) was evaluated, using the autochthonous Saccharomyces cerevisiae XG3 strain as a control. Microfermentations with pure culture of each strain were performed in duplicate and the basic parameters and major volatiles of wines were analysed following official methodology. The best strain within each species was selected using a quantification matrix including the relevant oenological characteristics. Results: The fermentative ability of non-Saccharomyces was lower than S. cerevisiae in all cases, but with differences among species. L. thermotolerans and T. delbrueckii showed higher fermentation rates than Starm. bacillaris, whereas Metschnikowia spp. presented a low fermentative power. At chemical level all non-Saccharomyces strains reduced the alcoholic content, the higher alcohols and the volatile acidity of wines and increased the content of glycerol, with differences among strains within a given species. T. delbrueckii and L. thermotolerans increased the total acidity of wines. The latter and Metschnikowia spp. strains produced lactic acid, which decreased the wine pH in the case of L. thermotolerans. According to their oenological traits the best rated strains of each species were Lt93, Td315, Mf278 and Sb474. In addition, the data obtained in pure fermentations were correlated to those chemical and aromatic compounds obtained with these non-Saccharomyces strains in sequential fermentations. Conclusions: Autochthonous strains of non-Saccharomyces yeast species contribute distinctive chemical characteristics to the wines. The correlations observed between wines fermented with the different non-Saccharomyces indigenous strains in pure and sequential fermentations suggest that their contribution to wine properties remains stable regardless of must composition or winemaking techniques.
Nowadays, there is a global trend towards differentiation and typicity of quality wine which is of great importance in commercial terms and as a generator of socio-economic value. However, in a context where wine regions are facing challenges such as the consequences of climate change, competition, market evolution or the development of new products, they require the use of new tools. In the last decades, different biological approaches as alternative strategies have been proposed to enhance complexity and aromatic profile of wines as well as to solve technical problems on analytical parameters such as ethanol or acidity [1, 2, 3]. Among these tools, indigenous non-Saccharomyces yeasts have been used to improve the quality and distinctiveness of wines associated with a given region [4, 5, 6].
In addition, non-Saccharomyces yeasts have shown other characteristics
of oenological relevance such as antimicrobial activity, bioprotective action,
and changes in the production of compounds such as ethanol, glycerol, or lactic
acid. In particular, Torulaspora delbrueckii has shown capacity to
increases the concentrations of desired aroma compounds of wine, to reduce the
final ethanol content, to act against spoilage yeasts and to decrease levels of
acetic acid and biogenic amines [1, 7, 8]. Lachancea thermotolerans is
characterized by a low production of volatile acidity; it shows a high production
of lactic acid, sometimes decreasing wine pH during fermentation, has relevant
potential to reduce ethanol content and improves the production of
2-phenylethanol and glycerol [3, 9, 10]. Other studies have shown that the presence
of Starmerella bacillaris increased the level of glycerol, and,
remarkably, reduced acetaldehyde and total SO
Most of these properties of non-Saccharomyces yeasts are strain-dependent; thus, the knowledge of the oenological potential of autochthonous strains is a valuable aid to their application. There is a great interest in the elaboration of wines with specific characteristics linked to a production area. This tendency has led to a rediscovery of the fermentation directed by native yeasts to obtain differentiated quality wines [4, 15]. In this sense, it is known that continued use of commercial strains can lead to the loss of complexity and the wines standardization, together with the decrease of yeast species diversity in the winemaking environments. Therefore, despite the availability of commercial cultures, the use of native non-Saccharomyces but also indigenous Saccharomyces cerevisiae strains can confer a specific aromatic imprint to the wine due to the metabolic interaction between S. cerevisiae/non-Saccharomyces strains in mixed starter cultures [9, 16, 17, 18, 19] contributing uniqueness to the wines in preserving the regional character from a given territory as a biogeographic or microbial terroir [4, 5, 6, 20, 21]. However, not all native strains show the desired oenological aptitudes. In this context, it is necessary to search for the oenological aptitudes of a wide range of strains. This study evaluated the potential of 29 different autochthonous strains within Metschnikowia spp., L. thermotolerans, T. delbrueckii and Starm. bacillaris species from the yeast culture collection maintained at Estación de Viticultura e Enoloxía de Galicia (EVEGA-AGACAL). Each strain was evaluated in pure fermentations and the results of the fermentative ability, and the chemical composition and aromatic profile of the resulting wines were used to select the best rated strains from each species. In addition, the results were correlated with those obtained in sequential fermentations of the selected strains with the autochthonous strain S. cerevisiae XG3 and with the commercial strain EC1118 to confirm the contribution of a given strain to wines.
Yeast strains used in this study were obtained from the yeast culture collection maintained at EVEGA-AGACAL. The autochthonous yeast strains were isolated from grapes and musts from organic and conventional vineyards in Galicia over several years, identified as explained in [22] and stored at EVEGA. Within this yeast collection, 29 non-Saccharomyces strains belonging to Metschnikowia spp. (10 strains), Starm. bacillaris (10 strains), L. thermotolerans (6 strains) and T. delbrueckii (3 strains), were chosen due to the oenological interest of these species in recent literature [2, 3, 23, 24]. Metschnikowia pulcherrima and Metschnikowia fructicola are two genetically very close species with a very similar oenological potential that are difficult to identify, so they have been considered in the same group [13]. Depending on the availability of indigenous strains isolated from each of the relevant species mentioned above, between 3 and 10 isolates were evaluated (Table 1). The use of strains isolated from different Denominations of Origin (DOs) and from different plots and cultivation systems within the same species allowed to explore different oenological behaviours to select the most appropriate ones. In addition, the S. cerevisiae XG3 strain, previously tested and selected at EVEGA, was used as a control [25, 26].
| Species | Strain | Denomination of origin | Year | Grapevine variety | Production system | Fermentation stage |
| Saccharomyces cerevisiae | ScXG3 | |||||
| Lachancea thermotolerans | Lt3 | Monterrei | 2015 | Treixadura | Organic | Final |
| Lt18 | Monterrei | 2015 | Treixadura | Organic | Initial | |
| Lt93 | Monterrei | 2015 | Mencía | Organic | Initial | |
| Lt132 | Monterrei | 2015 | Mencía | Organic | Initial | |
| Lt262 | Ribeiro | 2014 | Brancellao | Organic | Initial | |
| Lt205 | Monterrei | 2013 | Treixadura | Organic | Final | |
| Torulaspora delbrueckii | Td336 | Ribeira Sacra | 2015 | Mencía | Conventional | Must |
| Tdm5 | Rías Baixas | 2013 | Albariño | Organic | Must | |
| Td315 | Ribeiro | 2014 | Treixadura | Conventional | Final | |
| Starmerella bacillaris | Sb295 | Ribeira Sacra | 2015 | Mencía | Organic | Must |
| Sb306 | Ribeira Sacra | 2015 | Mencía | Organic | Must | |
| Sb326 | Ribeira Sacra | 2015 | Mencía | Organic | Initial | |
| Sb333 | Ribeira Sacra | 2015 | Mencía | Conventional | Must | |
| Sb404 | Rías Baixas | 2015 | Albariño | Organic | Initial | |
| Sb405 | Rías Baixas | 2015 | Albariño | Organic | Initial | |
| Sb304 | Ribeiro | 2014 | Treixadura | Conventional | Final | |
| Sb472 | Rías Baixas | 2015 | Treixadura | Organic | Initial | |
| Sb474 | Rías Baixas | 2015 | Treixadura | Organic | Final | |
| Sb494 | Rías Baixas | 2015 | Treixadura | Organic | Initial | |
| Metschnikowia fructicola | Mf278 | Ribeiro | 2015 | Treixadura | Conventional | Initial |
| Metschnikowia pulcherrima | Mp114 | Monterrei | 2015 | Mencía | Organic | Initial |
| Mp131 | Monterrei | 2015 | Mencía | Organic | Initial | |
| Mp176 | Ribeiro | 2015 | Treixadura | Organic | Initial | |
| Mp193 | Ribeiro | 2015 | Brancellao | Organic | Initial | |
| Mp205 | Ribeiro | 2015 | Brancellao | Organic | Initial | |
| Mp294 | Ribeira Sacra | 2015 | Mencía | Organic | Must | |
| Mp325 | Ribeira Sacra | 2015 | Mencía | Organic | Initial | |
| Mp385 | Rías Baixas | 2015 | Albariño | Organic | Initial | |
| Mp468 | Rías Baixas | 2015 | Treixadura | Organic | Must |
The original culture of each yeast strain was streaked on WL nutrient agar
medium to isolate single colonies. Then, a single colony was used for
pre-inoculum growth (for 24 h at 28 °C) with the aim of obtaining
sufficient biomass of fresh growth. The number of cells in the preinocula was
quantified by measurement of the optical density at 600 nm in a
spectrophotometer. Microfermentations were carried out in duplicate, using 75 mL
of pasteurized (10 min at 100 °C) white grape must in 100 mL
sterilized bottles closed with sterile-venting membrane screw caps. The strains
in pure culture were inoculated in flame-sterile conditions at a concentration of
5
Fermentations were carried out at a controlled temperature of 17
°C
Wines were analysed in the chemistry laboratory of EVEGA immediately after the
end of fermentation. General parameters (total acidity, volatile acidity, lactic
acid, malic acid, glucose + fructose, glycerol, alcohol content and pH) were
determined by Fourier transform infrared spectrometry (FTIR) using a Wine Scan
FT120 analyser (FOSS Electric, Barcelona, Spain) calibrated according to [27]. In
addition, the ethanol yield was calculated as the ethanol production (g·L
Volatile compounds including higher alcohols (methanol, propanol, isobutanol, 1-butanol, 2-methyl-1-butanol and 3-methyl-1-butanol), acetaldehyde and ethyl acetate were quantified in an Agilent model 7890A (Palo Alto, CA, USA) chromatograph with flame ionization detector (FID) as described by [28].
Significant differences between samples were tested by variance analysis (ANOVA)
and Tukey tests (p
Strain evaluation for each species was based on the weighted scoring method of
the different parameters among all wines made with all different strains (of all
species equally). The reference values established by the regulatory councils of
the Galician Denominations of Origin for quality white wines were used.
Therefore, the scores are comparable between strains of different species;
however, it is more appropriate to compare strains within a given species because
there are parameters such as the presence of reducing sugars that will penalise
species with lower fermentative capacity. A maximum value of ten points was
established for the positive range which was calculated by applying the following
function: f = 10
The mean value of the estimated number of cells in the pre-inoculum for all
strains was 10
 Fig. 1.
Fig. 1.Estimated number of cells in the pre-inoculum. Number of cells in preinocula of some strains checked by measurement of the optical density at 600 nm.
Fermentation kinetics is shown in Fig. 2. As expected, S. cerevisiae XG3 began to ferment within the first 2–3 days and showed a higher fermentation speed than non-Saccharomyces vinifications. The fermentative ability of autochthonous non-Saccharomyces strains in pure culture varied among species and strains, with L. thermotolerans and T. delbrueckii standing out for their high fermentation rates, but lower speed than S. cerevisiae, as reported by other works [8, 9, 10, 12, 32, 33]. Starm. bacillaris strains showed a lower fermentation ability than strains from L. thermotolerans and T. delbrueckii, although this species has been described as tolerant to relative high ethanol contents and can survive up to the middle-end phase of the fermentation process [12, 34]. Regarding Metschnikowia spp. strains, they exhibited low to moderate fermentative power, as this species is not able to tolerate ethanol concentrations over 4–5% (v/v), so it naturally disappears when this content of ethanol is exceeded during alcoholic fermentation [13]. M. fructicola and M. pulcherrima presented similar fermentation kinetics and fermentation yields over time. The poor or slower fermentative activity of non-Saccharomyces species confirmed the need to add a S. cerevisiae strain to successfully complete fermentation. However, the slower fermentation kinetics could be positive for a better retention of volatile compounds and for energy savings during the tanks cooling [3, 35]. Unsurprisingly, the control (pasteurised must sample w/o inoculum) showed no evidence of fermentation or contamination.
 Fig. 2.
Fig. 2.Fermentative yield of 30 autochthonous strains. Fermentative ability in pure culture of indigenous non-Saccharomyces strains and S. cerevisiae XG3. Lt, L. thermotolerans; Mf, M. fructicola; Mp, M. pulcherrima; Sb, Starm. bacillaris; Td, T. delbrueckii; Sc, S. cerevisiae.
Fermentations with different non-Saccharomyces strains influenced the chemical characteristics of wines. Basic chemical parameters and aromatic composition of wines are summarized in Tables 2,3, respectively. The results evidenced a significant effect of species and strains in almost all the parameters determined. T. delbrueckii and L. thermotolerans increased the total acidity and lowered the volatile acidity of wines compared with the control wines (fermented with S. cerevisiae XG3) (Fig. 3A). This ability has been widely reported for L. thermotolerans [36, 37, 38], but the results are less clear for T. delbrueckii [39]. To a lesser extent Metschnikowia spp. and Starm. bacillaris also decreased the volatile acidity as found in other studies [13, 36]. L. thermotolerans, especially Lt93 strain, also increased the lactic acid ratio, as well as Metschnikowia spp. but the M. pulcherrima strains did so to a lesser extent than L. thermotolerans. Accordingly, the pH was lower in the L. thermotolerans wines but not in all the wines fermented with Metschnikowia spp. Besides, L. thermotolerans strains reduced the amount of malic acid in the wine compared to S. cerevisiae. This ability is of special interest in warm regions to mitigate the acidity reduction in wines because of climate change [10, 29, 40].
| Strain | Total acidity | Volatile acidity | Lactic acid | Malic acid | Glucose + fructose | Glycerol | Alcohol content | Ethanol yield (g/g) | pH |
|---|---|---|---|---|---|---|---|---|---|
| (g |
(g |
(g·L |
(g·L |
(g·L |
(g·L |
(% vol) | |||
| ScXG3 | 6.8 |
0.60 |
0.1 |
3.5 |
0.2 |
3.8 |
10.05 |
0.42 |
3.66 |
| Lt3 | 7.9 |
0.39 |
1.7 |
2.6 |
6.1 |
5.6 |
9.12 |
0.39 |
3.58 |
| Lt18 | 7.4 |
0.38 |
0.8 |
2.85 |
20.3 |
5.6 |
8.08 |
0.38 |
3.58 |
| Lt93 | 8.0 |
0.43 |
2.6 |
2.3 |
0.8 |
6.1 |
9.02 |
0.38 |
3.53 |
| Lt132 | 7.6 |
0.40 |
1.5 |
2.5 |
9.8 |
5.5 |
9.12 |
0.40 |
3.65 |
| Lt262 | 6.9 |
0.36 |
0.5 |
3.0 |
6.5 |
4.9 |
8.71 |
0.38 |
3.68 |
| Lt205 | 7.3 |
0.36 |
0.9 |
2.8 |
0.6 |
5.2 |
8.75 |
0.37 |
3.58 |
| Lt | 7.5 |
0.39 |
1.3 |
2.6 |
7.4 |
5.5 |
8.8 |
0.38 |
3.60 |
| Td336 | 7.1 |
0.36 |
0.1 |
3.2 |
37.5 |
5.1 |
7.75 |
0.41 |
3.59 |
| Tdm5 | 7.7 |
0.61 |
0.1 |
3.9 |
2.5 |
3.3 |
9.25 |
0.39 |
3.63 |
| Td315 | 7.1 |
0.39 |
0.1 |
3.1 |
31.8 |
5.7 |
8.08 |
0.41 |
3.64 |
| Td | 7.3 |
0.45 |
0.1 |
3.4 |
23.9 |
4.7 |
8.4 |
0.40 |
3.62 |
| Sb295 | 6.8 |
0.50 |
0.1 |
3.5 |
55.5 |
16.7 |
6.60 |
0.39 |
3.85 |
| Sb306 | 6.9 |
0.49 |
0.1 |
3.4 |
44.0 |
14.5 |
7.34 |
0.40 |
3.76 |
| Sb326 | 6.5 |
0.41 |
0.1 |
3.4 |
67.4 |
16.7 |
6.22 |
0.41 |
3.82 |
| Sb333 | 6.9 |
0.62 |
0.1 |
3.3 |
47.7 |
16.1 |
6.92 |
0.39 |
3.78 |
| Sb404 | 6.2 |
0.54 |
0.1 |
3.0 |
29.8 |
13.4 |
7.84 |
0.39 |
3.71 |
| Sb405 | 6.5 |
0.37 |
0.1 |
3.6 |
66.9 |
16.6 |
6.25 |
0.41 |
3.85 |
| Sb304 | 7.1 |
0.50 |
0.1 |
3.2 |
0.3 |
7.0 |
9.70 |
0.41 |
3.58 |
| Sb472 | 6.5 |
0.53 |
0.1 |
3.1 |
41.1 |
15.2 |
7.38 |
0.40 |
3.77 |
| Sb474 | 6.8 |
0.39 |
0.1 |
3.5 |
65.9 |
17.0 |
6.35 |
0.41 |
3.92 |
| Sb494 | 6.5 |
0.49 |
0.1 |
3.3 |
47.8 |
15.9 |
6.91 |
0.39 |
3.77 |
| Sb | 6.6 |
0.48 |
0.1 |
3.3 |
46.6 |
14.9 |
7.1 |
0.40 |
3.78 |
| Mf278 | 6.7 |
0.42 |
0.6 |
1.6 |
78.5 |
11.6 |
4.16 |
0.30 |
3.69 |
| Mp114 | 6.8 |
0.50 |
0.3 |
2.0 |
86.5 |
12.4 |
3.61 |
0.28 |
3.68 |
| Mp131 | 6.3 |
0.52 |
0.3 |
1.6 |
83.5 |
12.9 |
3.71 |
0.28 |
3.72 |
| Mp176 | 7.2 |
0.73 |
0.3 |
2.7 |
97.5 |
10.1 |
3.65 |
0.31 |
3.78 |
| Mp193 | 6.8 |
0.46 |
0.4 |
2.0 |
83.5 |
10.8 |
4.10 |
0.31 |
3.64 |
| Mp205 | 7.1 |
0.47 |
0.4 |
2.3 |
88.0 |
11.5 |
3.94 |
0.31 |
3.69 |
| Mp294 | 6.6 |
0.44 |
0.2 |
2.8 |
92.5 |
11.3 |
3.66 |
0.31 |
3.64 |
| Mp325 | 7.2 |
0.44 |
0.6 |
1.8 |
95.5 |
13.2 |
3.52 |
0.30 |
3.71 |
| Mp385 | 6.5 |
0.58 |
0.4 |
2.0 |
83.8 |
11.3 |
3.99 |
0.30 |
3.63 |
| Mp468 | 7.5 |
0.54 |
0.4 |
2.2 |
88.0 |
11.8 |
3.68 |
0.29 |
3.69 |
| Mp | 6.8 |
0.51 |
0.4 |
2.1 |
87.7 |
11.6 |
3.8 |
0.30 |
3.68 |
| Sig. | *** | * | *** | *** | *** | *** | *** | *** | *** |
| Different letters in bold in the same column indicate significant
differences among species for that parameter according to Tukey’s test. Letters
in italics in the same column indicate significant differences among
strains within a given species at p | |||||||||
| Strain | 1-butanol | 2-methyl-1-butanol | 3-methyl-1-butanol | Isobutanol | Propanol | Methanol | Acetaldehyde | Ethyl acetate | |
|---|---|---|---|---|---|---|---|---|---|
| (mg·L |
(mg·L |
(mg·L |
(mg·L |
(mg·L |
(mg·L |
(mg·L |
(mg·L |
(mg·L | |
| ScXG3 | 185.0 |
3.4 |
24.1 |
110.7 |
20.3 |
26.8 |
22.5 |
8.5 |
4.5 |
| Lt3 | 136.5 |
2.9 |
23.7 |
73.2 |
19.1 |
24.8 |
25.5 |
24.5 |
4.7 |
| Lt18 | 162.5 |
4.9 |
27.4 |
87.7 |
20.8 |
29.8 |
21.2 |
23.8 |
3.2 |
| Lt93 | 159.4 |
2.7 |
24.8 |
88.2 |
21.4 |
28.1 |
25.5 |
33.5 |
4.3 |
| Lt132 | 116.0 |
5.9 |
14.1 |
49.4 |
22.1 |
32.2 |
23.5 |
18.5 |
8.7 |
| Lt262 | 154.9 |
4.6 |
25.6 |
85.4 |
17.3 |
28.3 |
22.0 |
25.5 |
4.8 |
| Lt205 | 162.0 |
4.7 |
25.0 |
84.0 |
15.2 |
33.0 |
21.0 |
15.0 |
4.0 |
| Lt | 148.0 |
4.3 |
23.4 |
77.9 |
19.3 |
29.3 |
23.1 |
23.5 |
4.9 |
| Td336 | 116.5 |
1.5 |
11.0 |
73.0 |
17.5 |
13.5 |
21.0 |
30.5 |
6.5 |
| Tdm5 | 131.0 |
1.5 |
13.0 |
58.0 |
17.0 |
32.5 |
21.0 |
16.5 |
6.0 |
| Td315 | 122.0 |
2.0 |
11.0 |
74.5 |
16.5 |
13.5 |
22.5 |
10.5 |
10.0 |
| Td | 123.2 |
1.7 |
11.7 |
68.5 |
17.0 |
19.8 |
21.5 |
19.2 |
7.5 |
| Sb295 | 80.5 |
- | 3.3 |
11.3 |
46.0 |
19.7 |
15.0 |
16.5 |
8.5 |
| Sb306 | 81.5 |
- | 3.9 |
12.8 |
39.2 |
25.6 |
24.5 |
27.5 |
11.5 |
| Sb326 | 43.0 |
- | 4.0 |
6.0 |
11.7 |
20.1 |
26.0 |
20.0 |
7.0 |
| Sb333 | 93.0 |
- | 4.6 |
11.7 |
46.1 |
31.3 |
22.0 |
32.5 |
12.0 |
| Sb404 | 77.5 |
- | 4.6 |
12.5 |
33.4 |
26.8 |
21.0 |
22.5 |
6.5 |
| Sb405 | 84.3 |
0.4 |
7.5 |
23.4 |
33.7 |
19.8 |
23.0 |
23.0 |
6.5 |
| Sb304 | 60.0 |
- | 4.1 |
9.2 |
21.2 |
24.9 |
23.5 |
17.5 |
8.5 |
| Sb472 | 61.0 |
- | 3.8 |
9.9 |
22.3 |
23.4 |
24.5 |
36.0 |
9.5 |
| Sb474 | 139.0 |
6.2 |
15.4 |
52.1 |
32.5 |
33.7 |
22.0 |
20.5 |
6.5 |
| Sb494 | 84.5 |
- | 10.1 |
14.6 |
38.6 |
26.5 |
20.0 |
22.0 |
10.0 |
| Sb | 81.02 |
3.3 |
6.1 |
16.4 |
33.1 |
25.2 |
22.1 |
23.7 |
8.6 |
| Mf278 | 110.2 |
0.9 |
8.4 |
43.9 |
47.0 |
9.9 |
18.0 |
21.5 |
12.0 |
| Mp114 | 116.5 |
1.0 |
7.5 |
50.0 |
51.0 |
8.0 |
19.5 |
10.5 |
40.5 |
| Mp131 | 109.0 |
1.0 |
8.0 |
42.0 |
49.0 |
10.0 |
20.5 |
10.5 |
201.0 |
| Mp176 | 104.5 |
0.0 |
8.3 |
34.9 |
46.6 |
14.7 |
21.0 |
13.0 |
330.0 |
| Mp193 | 122.0 |
0.0 |
6.5 |
51.5 |
54.5 |
9.0 |
20.5 |
8.5 |
55.0 |
| Mp205 | 104.0 |
0.0 |
7.0 |
38.0 |
49.5 |
8.5 |
29.5 |
8.5 |
45.5 |
| Mp294 | 117.0 |
0.0 |
7.0 |
39.5 |
58.5 |
11.5 |
33.5 |
13.0 |
89.5 |
| Mp325 | 99.0 |
1.0 |
7.5 |
41.0 |
42.0 |
7.0 |
19.0 |
10.0 |
36.0 |
| Mp385 | 159.0 |
- | 5.7 |
67.3 |
72.9 |
13.3 |
15.5 |
45.0 |
133.0 |
| Mp468 | 108.0 |
- | 4.2 |
36.2 |
64.5 |
3.4 |
12.0 |
26.0 |
67.0 |
| Mp | 114.9 |
0.5 |
6.9 |
44.4 |
53.5 |
9.5 |
20.9 |
16.6 |
100.9 |
| Sig. | *** | *** | *** | *** | *** | *** | ns | ns | *** |
| The values are mean | |||||||||
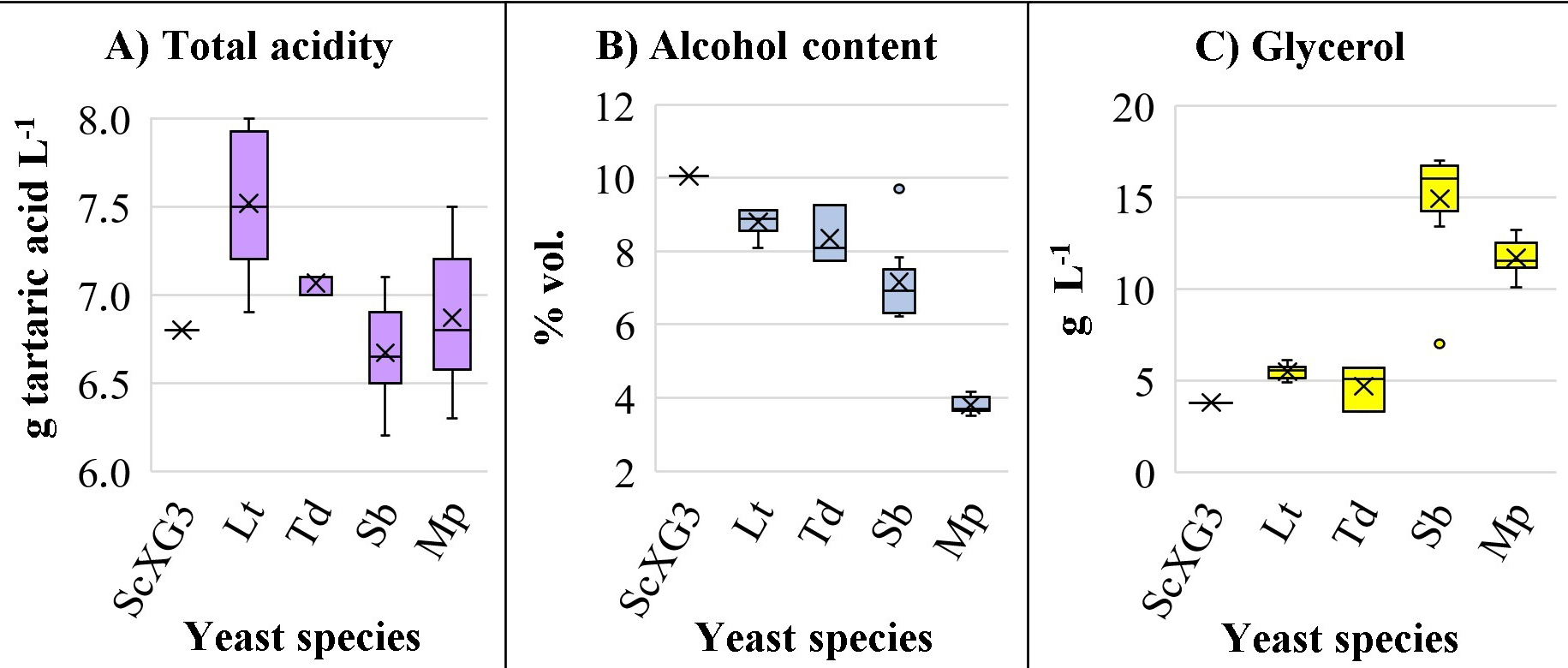 Fig. 3.
Fig. 3.Total acidity, alcohol content and glycerol. Box and whisker chart of three relevant basic chemical compounds: (A) alcohol content, (B) total acidity, and (C) glycerol of non-Saccharomyces strains compared to S. cerevisiae XG3.
Moreover, the alcoholic degree of non-Saccharomyces wines was lower
than control in all species and strains (Fig. 3B). For instance, a reduction of
1.25% v/v in the case of L. thermotolerans, 1.69% v/v with T.
delbrueckii, and 2.90% v/v with Starm. bacillaris was observed,
although in most cases there was residual sugars in the wines. Accordingly, the
wines from these species presented lower ethanol yields than control wines
obtained with ScXG3. Concerning to alcoholic strength and sugar consumption by
the different non-Saccharomyces in single fermentations, results are
consistent with the literature, but some strains such as Lt93 reached higher
ethanol yield (ethanol production (g·L
Finally, all non-Saccharomyces strains increased the concentration of glycerol in the wine, especially Starm. bacillaris and Metschnikowia spp. (Fig. 3C). Particularly, with some strains of Starm. bacillaris the content of glycerol was very high (17.0 in strain Sb474), as described by other authors [2, 11, 12, 36]. This compound contributes unctuousness, volume, slight sweetness and silky mouthfeel to wines [12, 44, 45].
Yeast strains modulate the aroma of wine during fermentation. In particular,
higher alcohols are the major compounds that also contribute to the generation of
secondary metabolites whose biosynthesis is species dependent but not always is
strain dependent [46]. Accordingly, all the yeast strains/species used in this
study influenced the fermentative aroma composition of wines. Other major
volatile compounds include acetaldehyde and ethyl acetate, both are undesirable
at high concentrations imparting aroma like pungent and nail-polish,
respectively. However, below 0.5 mg·L
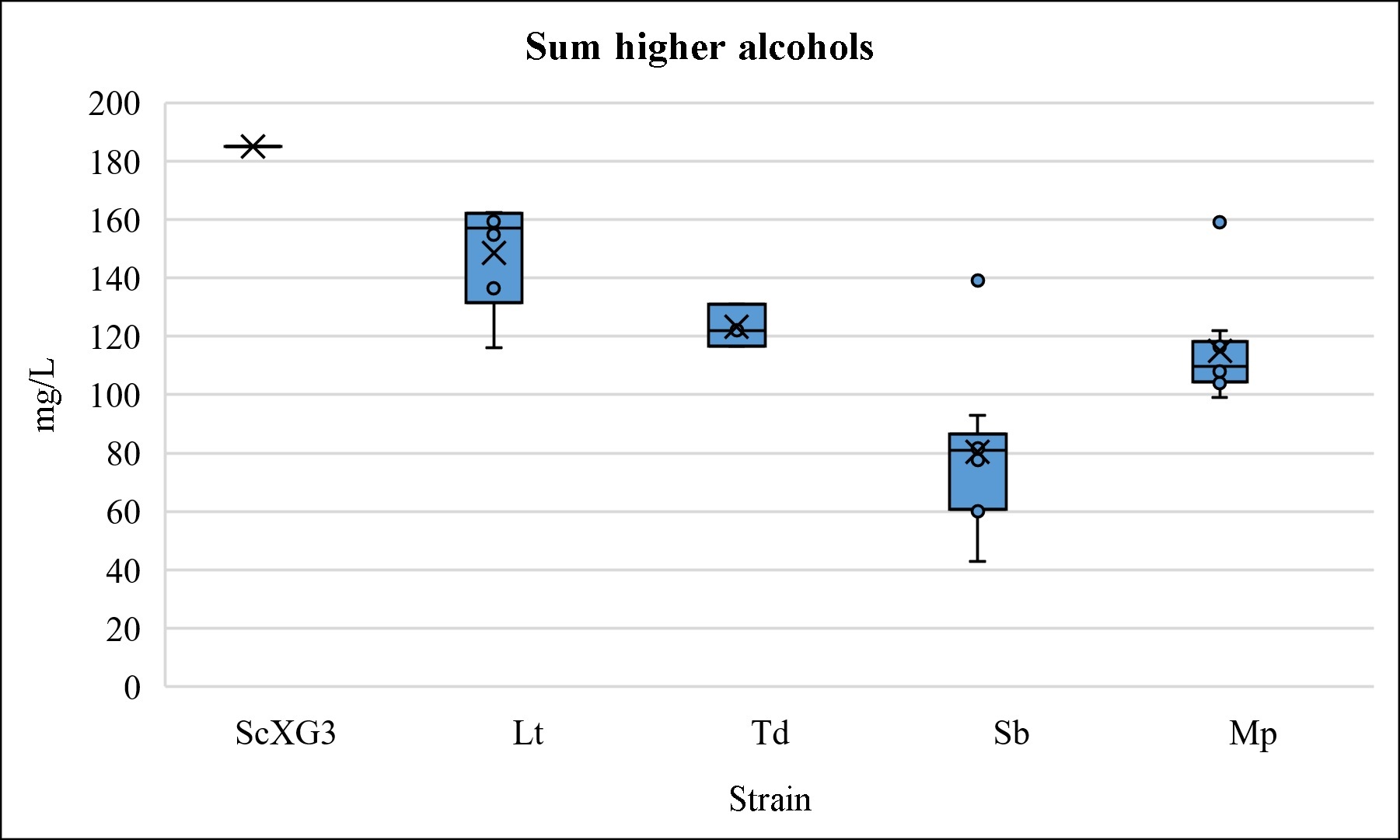 Fig. 4.
Fig. 4.Higher alcohols content. Box and whisker chart of higher alcohols content comparing the non-Saccharomyces strains to S. cerevisiae XG3. The sum of higher alcohols included 1-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol, isobutanol (2-methyl-1-propanol) and propanol.
On the other hand, the high levels of ethyl acetate in wines fermented with some strains of Metschnikowia spp. are noteworthy. However, other strains obtained very low non-altering concentrations of this compound, which reinforces the argument that its production is strain-dependent; therefore, the strain selection is very important [23, 36].
The data from the chemical analyses (Tables 2,3) of the wines were included
in a quantification matrix (Table 4), evaluating each of the desired oenological
characteristics. Lt93 was characterised by high total acidity (+1.2 g·L
| Parameter | ScXG3 |
Lt3 |
Lt18 |
Lt93 |
Lt132 |
Lt262 |
Lt205 |
Td336 |
Tdm5 |
Td315 |
Sb295 |
Sb306 |
Sb326 |
Sb333 |
Sb404 |
Sb405 |
Sb304 |
Sb472 |
Sb474 |
Sb494 |
Mf278 |
Mp114 |
Mp131 |
Mp176 |
Mp193 |
Mp205 |
Mp294 |
Mp325 |
Mp385 |
Mp468 |
Weighting |
| Higher alcohols | 10.0 | 7.4 | 8.8 | 8.6 | 6.3 | 8.4 | 8.8 | 6.3 | 7.1 | 6.6 | 4.4 | 4.4 | 2.3 | 5.0 | 4.2 | 4.6 | 3.2 | 3.3 | 7.5 | 4.6 | 6.0 | 6.3 | 5.9 | 5.6 | 6.6 | 5.6 | 6.3 | 5.4 | 8.6 | 5.8 | 1.1 |
| Acetaldehyde | 8.1 | 4.6 | 4.7 | 2.6 | 5.9 | 4.3 | 6.7 | 3.2 | 6.3 | 7.7 | 6.3 | 3.9 | 5.6 | 2.8 | 5.0 | 4.9 | 6.1 | 2.0 | 5.4 | 5.1 | 5.2 | 7.7 | 7.7 | 7.1 | 8.1 | 8.1 | 7.1 | 7.8 | 0.0 | 4.2 | 1 |
| Methanol | 6.7 | 7.6 | 6.3 | 7.6 | 7.0 | 6.6 | 6.3 | 6.3 | 6.3 | 6.7 | 4.5 | 7.3 | 7.8 | 6.6 | 6.3 | 6.9 | 7.0 | 7.3 | 6.6 | 6.0 | 5.4 | 5.8 | 6.1 | 6.3 | 6.1 | 8.8 | 10.0 | 5.7 | 4.6 | 3.6 | 0.8 |
| Ethyl acetate | 9.6 | 9.6 | 9.7 | 9.6 | 9.3 | 9.6 | 9.7 | 9.5 | 9.5 | 9.2 | 9.3 | 9.0 | 9.4 | 9.0 | 9.5 | 9.5 | 9.3 | 9.2 | 9.5 | 9.2 | 9.0 | 6.6 | –6.8 | –17.5 | 5.4 | 6.2 | 2.5 | 7.0 | –1.1 | 4.4 | 1.1 |
| Total acidity (g tart L |
8.5 | 9.9 | 9.3 | 10.0 | 9.5 | 8.6 | 9.1 | 8.9 | 9.6 | 8.9 | 8.5 | 8.6 | 8.1 | 8.6 | 7.8 | 8.1 | 8.9 | 8.1 | 8.5 | 8.1 | 8.4 | 8.5 | 7.9 | 9.0 | 8.5 | 8.9 | 8.3 | 9.0 | 8.1 | 9.4 | 1.2 |
| Volatile acidity (g acetic L |
2.0 | 6.7 | 6.8 | 6.1 | 6.5 | 7.1 | 7.1 | 7.1 | 1.6 | 6.7 | 5.2 | 5.3 | 6.4 | 1.3 | 4.6 | 6.9 | 5.2 | 4.7 | 6.7 | 5.3 | 6.2 | 5.2 | 4.9 | –0.5 | 5.7 | 5.6 | 6.0 | 6.0 | 4.1 | 4.6 | 1.3 |
| Lactic acid | 0.4 | 6.5 | 3.1 | 10.0 | 5.8 | 1.9 | 3.5 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 2.3 | 1.2 | 1.2 | 1.2 | 1.5 | 1.5 | 0.8 | 2.3 | 1.5 | 0.4 | 1 |
| Malic acid | 9.0 | 6.7 | 7.3 | 5.9 | 6.4 | 7.7 | 7.2 | 8.2 | 10.0 | 7.9 | 9.0 | 8.7 | 8.7 | 8.5 | 7.7 | 9.2 | 8.2 | 7.9 | 9.0 | 8.5 | 4.1 | 5.1 | 4.1 | 6.9 | 5.1 | 5.9 | 7.2 | 4.6 | 5.1 | 5.6 | 1 |
| Reducing sugars* | 10.0 | 9.4 | 7.9 | 9.9 | 9.0 | 9.3 | 9.9 | 6.2 | 9.7 | 6.7 | 4.3 | 5.5 | 3.1 | 5.1 | 6.9 | 3.1 | 10.0 | 5.8 | 3.2 | 5.1 | 1.9 | 1.1 | 1.4 | 0.0 | 1.4 | 1.0 | 0.5 | 0.2 | 1.4 | 1.0 | 1.2 |
| Glycerol | 2.2 | 3.3 | 3.3 | 3.6 | 3.2 | 2.9 | 3.1 | 3.0 | 1.9 | 3.4 | 9.8 | 8.5 | 9.8 | 9.5 | 7.9 | 9.8 | 4.1 | 8.9 | 10.0 | 9.4 | 6.8 | 7.3 | 7.6 | 5.9 | 6.4 | 6.8 | 6.6 | 7.8 | 6.6 | 6.9 | 1.1 |
| Alcoholic strength (%vol.) | 10.0 | 9.1 | 8.0 | 9.0 | 9.1 | 8.7 | 8.7 | 7.7 | 9.2 | 8.0 | 6.6 | 7.3 | 6.2 | 6.9 | 7.8 | 6.2 | 9.7 | 7.3 | 6.3 | 6.9 | 4.1 | 3.6 | 3.7 | 3.6 | 4.1 | 3.9 | 3.6 | 3.5 | 4.0 | 3.7 | 1 |
| pH | 8.1 | 8.3 | 8.3 | 8.4 | 8.1 | 8.1 | 8.3 | 8.3 | 8.2 | 8.2 | 7.7 | 7.9 | 7.8 | 7.9 | 8.0 | 7.7 | 8.3 | 7.9 | 7.6 | 7.9 | 8.0 | 8.1 | 8.0 | 7.9 | 8.2 | 8.0 | 8.2 | 8.0 | 8.2 | 8.0 | 1 |
| Sum | 84.6 | 88.9 | 83.5 | 91.3 | 86.1 | 83.1 | 88.2 | 74.9 | 79.9 | 80.3 | 75.9 | 76.9 | 75.5 | 71.5 | 76.0 | 77.3 | 80.3 | 73.0 | 80.6 | 76.3 | 67.5 | 66.4 | 51.6 | 35.5 | 67.1 | 70.3 | 67.1 | 67.2 | 51.2 | 57.7 | |
| Weighted score | 89.8 | 95.3 | 89.9 | 97.8 | 92.2 | 89.6 | 95.0 | 80.7 | 84.9 | 86.0 | 81.4 | 82.0 | 80.3 | 75.6 | 81.2 | 82.6 | 85.9 | 77.9 | 86.4 | 81.6 | 72.6 | 70.8 | 54.4 | 35.3 | 71.4 | 74.1 | 70.2 | 71.7 | 54.8 | 62.2 | |
| Values are the average of two fermentations. * Glucose + fructose. ** Weighting factor used for each parameter according to its relative importance on the total wine quality. | |||||||||||||||||||||||||||||||
According to these values, the best rated strains within L. thermotolerans, T. delbrueckii, Starm. bacillaris and Metschnikowia spp. species were Lt93, Td315, Sb474 and Mf278, respectively. These strains were evaluated in sequential fermentation with the autochthonous S. cerevisiae strain XG3 [29] and with the commercial strain S. cerevisiae EC1118 [30].
The PCA using all chemical compounds analysed allowed a clear separation of wines elaborated with different strains of non-Saccharomyces species in pure culture (Fig. 5A). The first two components, PC1 and PC2, explained 60.5% of the variance. L. thermotolerans strains were located in the positive first quadrant, characterized by higher concentrations of 1-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol, total acidity and lactic acid. In contrast, wines made with Starm. bacillaris were plotted on the negative side of PCA (except for the Sb474 and Sb304 strains), with a high content of glycerol. Torulaspora wines appeared close to the centre, between Lt and Sb strains, due to their higher alcohol content. In the opposite, on the negative side of PC2 and in the positive side of PC1 Metschnikowia wines were characterized by higher content of sugars, ethyl acetate and isobutanol.
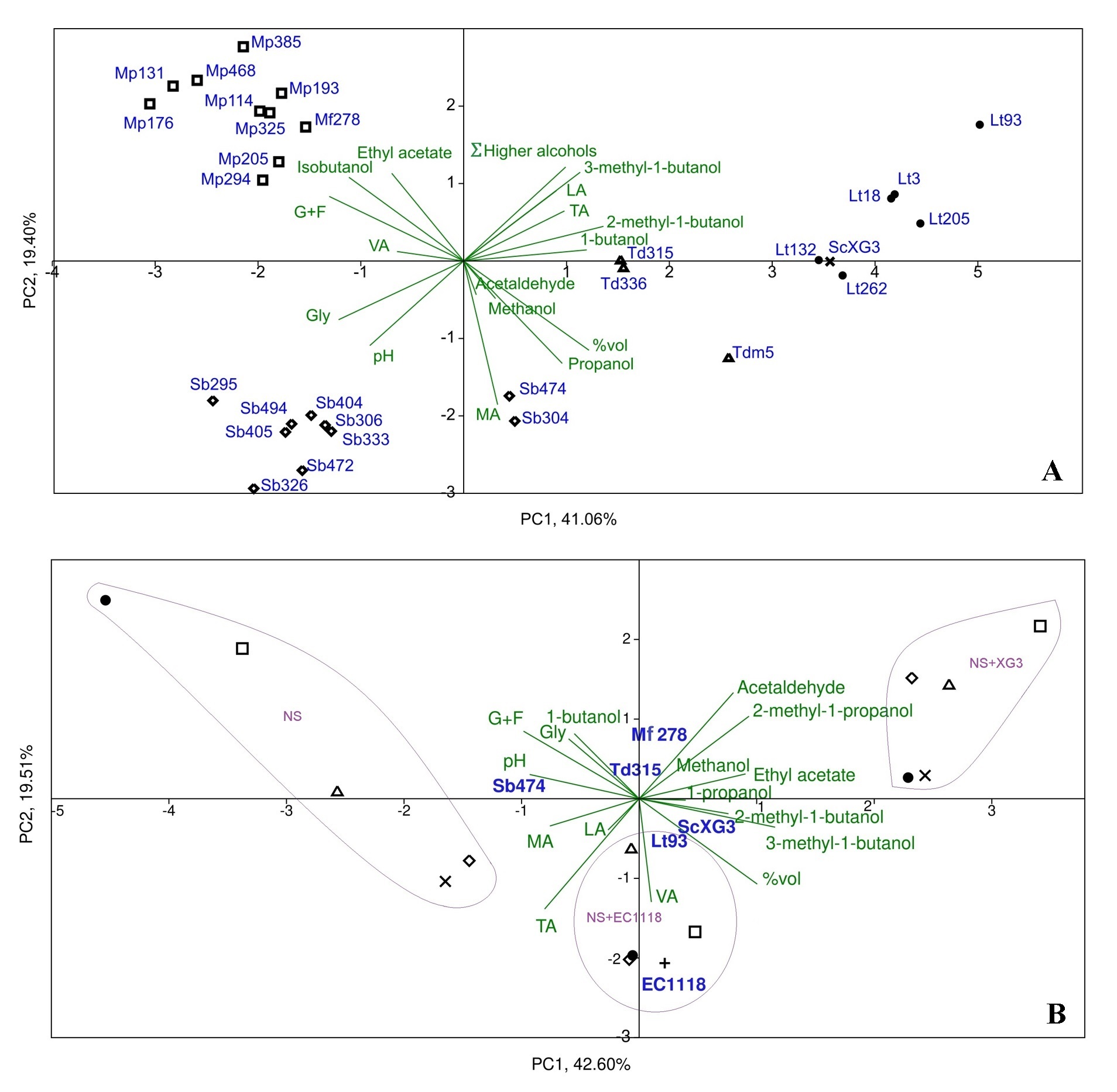 Fig. 5.
Fig. 5.Principal component analysis (PCA). (A) Pure cultures of autochthonous non-Saccharomyces strains: Metschnikowia spp. (square), L. thermotolerans (dot), Starm. bacillaris (diamond) and T. delbrueckii (triangle). Chemical parameter: %vol., alcohol content; G+F, reducing sugars (glucose + fructose); Gly, glycerol; LA, lactic acid; MA, malic acid; TA, total acidity; VA, volatile acidity. (B) Pure cultures of autochthonous non-Saccharomyces strains (NS), sequential fermentation with S. cerevisiae autochthonous XG3 (x-cross) and commercial EC1118 (plus). Biplot for the first two components (PC) for basic parameters and volatile compounds: ScXG3-fermentation with S. cerevisiae ScXG3; Lt93, Sb474, Td315 and Mf278-fermentations with each of these non-Saccharomyces strains and sequential inoculation with S. cerevisiae ScXG3 (NS+XG3) and S. cerevisiae EC1118 (NS+EC1118).
In addition, the results obtained with the best rated strains in pure fermentations were compared with those obtained with the same non-Saccharomyces strains in sequential fermentations. PCA results also shown a proper separation of wines elaborated with non-Saccharomyces strains in pure culture and in sequential fermentations (Fig. 5B). In this case, the first two components, PC1 and PC2, explained 62.1% of the variance. Wines made with non-Saccharomyces in pure culture (NS) were in left side. On the opposite side were grouped the wines produced by sequential fermentation with ScXG3. However, wines fermented in sequential fermentation with the commercial ScEC1118 strain (NS+EC1118) were plotted in the middle of the PCA. In addition, strains from the sequential fermentations, especially those from the NS+EC1118 group, clustered more closely than those from the pure culture, indicating greater homogeneity due to the influence of S. cerevisiae strains on the resulting wines while retaining their differentiation. Some authors have proposed different mechanisms in the relationships between S. cerevisiae and non-Saccharomyces [18, 51, 52]. In this sense, the NS+XG3 group showed the highest concentration of volatile compounds, which have confirmed that native strains can improve the chemical and sensory characteristics, and differentiation of wines in accordance with numerous recent studies [3, 30, 53, 54, 55]. In contrast, compounds accounting for acidity in the biplot are concentrated in the area of NS+EC1118 wines. On the other hand, some chemical compounds such as glycerol do not point to the species that produce the highest concentration in the biplot, which indicates that they do not depend on the type of fermentation but rather depends on the species and strain.
A correlation (Pearson) study (Table 5) was performed to show possible
correlations between chemical compounds, strains, and fermentation groups (pure
and sequential cultures). Regarding correlations between strains in single
fermentations, the results showed some differences. As expected, the correlations
between S. cerevisiae control strains in pure culture fermentations
showed high r-values with each other (r
| Basic compounds | Mayor volatile compounds | All compounds | ||||||||||||
| r overall | NS+XG3 | NS+EC1118 | Pure | r overall | NS+XG3 | NS+EC1118 | Pure | r overall | ScXG3 | ScXG3* | EC1118 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS+XG3 | 0.01119 | 0.90839 | NS+XG3 | 0.00097 | 0.00053 | ScXG3 | 3.60E-11 | 3.64E-08 | ||||||
| NS+EC1118 | 0.49859 | 0.63378 | NS+EC1118 | 0.93621 | 0.00948 | ScXG3+ | 0.97971 | 2.96E-07 | ||||||
| Pure | 0.01879 | 0.10017 | Pure | 0.54862 | 0.56474 | EC1118 | 0.99466 | 0.99028 | ||||||
| r G+F | NS+XG3 | NS+EC1118 | Pure | r 1-propanol | NS+XG3 | Pure | r overall | Lt93 | Lt93+XG3 | Lt93+EC1118 | ||||
| NS+XG3 | 0.0669 | 0.86208 | NS+XG3 | 0.22280 | Lt93 | 1.95E-08 | 0.00023 | |||||||
| NS+EC1118 | 0.85186 | 0.71677 | Pure | 0.66277 | Lt93+XG3 | 0.95816 | 0.00281 | |||||||
| Pure | 0.10854 | –0.22434 | Lt93+EC1118 | 0.93407 | 0.89319 | |||||||||
| r TA | NS+XG3 | NS+EC1118 | Pure | r 2-methyl-1-propanol | NS+XG3 | NS+EC1118 | Pure | r overall | Td315 | Td315+XG3 | Td315+EC1118 | |||
| NS+XG3 | 0.57074 | 0.67839 | NS+XG3 | 0.05638 | 0.80942 | Td315 | 1.64E-05 | 1.27E-05 | ||||||
| NS+EC1118 | –0.34405 | 0.14339 | NS+EC1118 | 0.86806 | 0.86528 | Td315+XG3 | 0.87850 | 0.00084 | ||||||
| Pure | –0.2554 | 0.75108 | Pure | 0.15025 | 0.10601 | Td315+EC1118 | 0.97139 | 0.92935 | ||||||
| r VA | NS+XG3 | NS+EC1118 | Pure | r 3-methyl-1-butanol | NS+XG3 | NS+EC1118 | Pure | r overall | Sb474 | Sb474+XG3 | Sb474+EC1118 | |||
| NS+XG3 | 0.04615 | 0.45604 | NS+XG3 | 0.04109 | 0.04054 | Sb474 | 0.02962 | 0.17303 | ||||||
| NS+EC1118 | 0.88474 | 0.65738 | NS+EC1118 | 0.89342 | 0.06032 | Sb474+XG3 | 0.56086 | 0.00186 | ||||||
| Pure | –0.44208 | –0.27251 | Pure | -0.89438 | -0.86188 | Sb474+EC1118 | 0.49744 | 0.90720 | ||||||
| r Gly | NS+XG3 | NS+EC1118 | Pure | r Acetaldehyde | NS+XG3 | NS+EC1118 | Pure | r overall | Mf278 | Mf278+XG3 | Mf278+EC1118 | |||
| NS+XG3 | 0.00219 | 0.00969 | NS+XG3 | 0.33377 | 0.42275 | Mf278 | 0.08303 | 0.38415 | ||||||
| NS+EC1118 | 0.98503 | 0.00230 | NS+EC1118 | 0.55288 | 0.00079 | Mf278+XG3 | 0.46191 | 0.00484 | ||||||
| Pure | 0.95960 | 0.98456 | Pure | 0.47147 | 0.99246 | Mf278+EC1118 | 0.33108 | 0.87118 | ||||||
| r MA | NS+XG3 | Pure | r Ethyl acetate | NS+XG3 | Pure | r overall | ScXG3 | Lt93 | Td315 | Sb474 | Mf278 | |||
| ScXG3 | 9.51E-09 | 2.68E-06 | 0.03651 | 0.21111 | ||||||||||
| NS+XG3 | 0.17011 | NS+XG3 | 0.04923 | Lt93 | 0.95456 | 3.03E-05 | 0.03232 | 0.19956 | ||||||
| Td315 | 0.89596 | 0.85019 | 0.00033 | 0.00457 | ||||||||||
| Pure | 0.72009 | Pure | 0.87960 | Sb474 | 0.52564 | 0.53608 | 0.78386 | 5.51E-07 | ||||||
| Mf278 | 0.33056 | 0.33860 | 0.66932 | 0.91768 | ||||||||||
| Chemical parameter: G+F, reducing sugars (glucose + fructose); Gly, glycerol; MA, malic acid; TA, total acidity; VA, volatile acidity. ScXG3: control in pure culture, ScXG3*: control in sequential fermentation. The p-value (significance) is shown above the diagonal and the r-value (Pearson’s correlation factor) is shown below the diagonal. NS (autochthonous non-Saccharomyces). | ||||||||||||||
On the other hand, the main correlations with significant differences
(p
The results of the PCA and Pearson test showed that the different non-Saccharomyces strains influenced the chemical profile of wines, through the formation of different compounds. Sequential inoculation of strain Mf278 increased the production of positive aroma compounds with respect to the indigenous control strain S. cerevisiae ScXG3 alone; it should be taken into consideration that ScXG3 has been selected in previous studies for its optimal winemaking behaviour [26, 56, 57]. Differences could be due to the fermentation process, and it should be considered for the use of these strains in future studies. T. delbrueckii and Starm. bacillaris strains were the worst qualified; however, several authors obtained better wines than their controls made with pure S. cerevisiae following a co-inoculated strategy with these species and S. cerevisiae [1, 18, 42, 58]. In fact, there are already commercial strains of T. delbrueckii and to a lesser extent of Starm. bacillaris that can be used due to their desirable oenological characteristics such as increased glycerol concentration, acidity, low ethanol and esters production [12, 59].
All these results are consistent with those found in the previous literature and allow us to affirm that some indigenous strains of non-Saccharomyces yeasts, used by sequential inoculation, can contribute to the typicity and differentiation of wines and even increase the contribution of aromatic compounds with positive sensory evaluation [1, 3, 55]. In this sense, the differentiation and correlations obtained between the different strains and fermentations show that the choice of yeast strain affects the chemical profile of the wines, but certain distinctive parameters are maintained in sequential fermentation.
Therefore, we suggest further research, with different strains and even by applying more than one non-Saccharomyces together (in multistarter yeast inoculum or sequentially before inoculation of S. cerevisiae) which has been proposed to compensate the shortage of one non-Saccharomyces species and further enhance the wine quality [18]. This will make it possible to evaluate the positive synergistic effects on wine quality and typicity, the interaction between species and ultimately increase the knowledge of interspecific relationships. Accordingly, combinations of T. delbrueckii, Hanseniaspora vineae, Hanseniaspora uvarum, M. pulcherrima, Zygosaccharomyces bailii or L. thermotolerans were reported as a multistarter or co-inoculation fermentations improving the chemical and aromatic compositions of wines fermented only with S. cerevisiae [7, 18, 39, 60].
Autochthonous strains of non-Saccharomyces yeast species contribute distinctive chemical characteristics to the wines. L. thermotolerans and T. delbrueckii strains influence acidity or alcohol content; Starm. bacillaris and Metschnikowia spp. increase glycerol concentration. The correlations observed between wines fermented with the different non-Saccharomyces indigenous strains in pure and sequential fermentations suggest that their contribution to wines properties remains stable regardless must composition or winemaking techniques. These results can help winemakers in their choice of species and strains.
DC and PB designed the research study. DC and PB performed the research. DC designed and developed the chemical analysis (gas chromatography). DC analysed the data. DC and PB wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
The authors thank to EVEGA Chemical Laboratory for the analysis of wine basic parameters. They are also grateful to Noemi Neira and Eva Rabuñal for their invaluable technical assistance in the laboratory.
This work was supported by the National Institute of Agricultural and Food Research and Technology [RTA2012–00021-C03–01], co-financed with ERDF funds. David Castrillo thanks the INIA and European Social Fund for his FPI pre-doctoral contract.
The authors declare no conflict of interest.
