†These authors contributed equally.
Academic Editor: Yasuhito Shimada
Background: Halogenated aromatic compounds are more resistant to microbial degradation than non-halogenated aromatic compounds. Microbial degradation of sodium benzoate in the presence of sodium 3-chlorobenzoate is of interest. The ability to degrade aromatic compounds is largely determined by the substrate specificity of the first enzyme that initiates degradation, namely, benzoate 1,2-dioxygenase for benzoate degradation, and 3-chlorobenzoate 1,2-dioxygenase for 3-chlorobenzoate degradation. In this study, the perspective of immobilized cells of Rhodococcus opacus 1CP actinobacterium for degradation of benzoate and 3-chlorobenzoate was explored. Methods: The biosensor approach (a membrane microbial sensor based on immobilized cells of Rhodococcus opacus 1CP and the Clark-type oxygen electrode as a transducer) was applied to evaluate the actinobacterial cells’ responses to benzoate and 3-chlorobenzoate in the absence of both enzymes, benzoate 1,2-dioxygenase and 3-chlorobenzoate 1,2-dioxygenase, or in the presence of one of the said enzymes. Results: Data obtained show that 1CP actinobacterium possessed a constitutive system for the transport of benzoate and 3-chlorobenzoate into culture cells. The affinity of the transport system for benzoate was higher than that for 3-chlorobenzoate. Moreover, adaptation to one substrate did not preclude the use of the second substrate. Probably, porins facilitated the penetration of benzoate and 3-chlorobenzoate into 1CP cells. Analyzing V vs. S dependencies, negative cooperativity was found, when benzoate 1,2-dioxygenase bound substrate (3-chlorobenzoate), while positive cooperativity was determined at benzoate binding. The observed difference could be associated with the presence of at least two systems of 3-chlorobenzoate transport into actinobacterial cells and allosteric interaction of active sites of benzoate 1,2-dioxygenase in the presence of 3-chlorobenzoate. Conclusions: The membrane microbial sensor based on immobilized Rhodococcus opacus 1CP cells could be useful as a perspective tool for comparative evaluation of enzymes of complex structure such as benzoate- and 3-chlorobenzoate 1,2-dioxygenase.
Salts of benzoic acid (benzoates) and chlorobenzoic acids (CBAs) enter the biosphere because of human activity in a household environment as well as during vital activity of living organisms and different reactions that occur in nature [1, 2, 3, 4, 5, 6, 7, 8, 9]. The harmfulness of benzoates is still discussed, but most of chlorinated aromatic compounds are toxic. It is known, that halogenated aromatic compounds are usually more resistant to microbial degradation than non-halogenated aromatic compounds. With an increase in the number of substituents in the ring, this resistance enhances. So, new remediation methods, including biological techniques, are being developed [2].
For bioremediation of polluted soil, two approaches such as activation of detoxifying ability of soil microflora and introduction of xenobiotics-degrading strains are used. Many species of the genus Pseudomonas and Rhodococcus can degrade a wide spectrum of xenobiotics [4, 10, 11, 12, 13]. The search for and introduction of new promising strains able to degrade various pollutants is urgent. For example, the Gram-positive actinobacterium Rhodococcus opacus 1CP can be used to detoxify compounds such as (chloro)phenols, (chloro/methyl) benzoates including 3-chlorobenzoate (3CBA) [14, 15]. To use a culture as a degrader of xenobiotics, it is important to know about the influence of xenobiotics on the microorganism and the activity of the enzymes involved in xenobiotic metabolism.
The ability to degrade (halogen-substituted) aromatic compounds is mainly determined by the substrate specificity of the first enzyme that initiates degradation. Benzoate 1,2-dioxygenase initiates benzoate metabolism (Fig. 1). It is considered that benzoate 1,2-dioxygenase is unable to perform an oxygenase attack on the 3-chlorobenzoate ring. Regarding the microbial degradation of 3CBA, there are several ways of degradation of this xenobiotic in bacterial cells. One of the ways is oxidation of the aromatic ring by 3-chlorobenzoate 1,2-dioxygenase enzyme [16] as shown in Fig. 1.
 Fig. 1.
Fig. 1.Comparison of structural formulas of BA, 3CBA and mandelate; the first steps of BA and 3CBA metabolism in R. opacus 1CP.
Benzoate 1,2-dioxygenase (BDO) is a two-component enzyme of complex structure that catalyzes the oxygen-dependent reaction [17]. 3-chlorobenzoate 1,2-dioxygenase (3CBDO) is the enzyme of peripheral metabolism that initiates the degradation of 3CBA. This enzyme catalyzes the reaction with the involvement of oxygen. Although activity of stable soluble enzymes may be determined in cell-free extracts of culture, activities of BDO and 3CBDO cannot be determined in cell-free extracts because the enzymes are destroyed during preparation of cell-free extracts [18]. For that reason, BDO and 3CBDO activities are determined in whole cells by a change of oxygen consumption by cells in the presence of the substrate of the enzyme under study [19, 20, 21].
The biosensor on the basis of bacterial cells is a helpful convenient tool for rapid assessment of the substrate specificity and activity of the enzyme initiating the pathway in this bacterium [22]. A whole-cell biosensor with the Clark-type oxygen electrode as a transducer can be used for estimation of a change in oxygen consumption by microbial cells under the action of a substrate. In our previous study, BDO and 3CBDO activities in R. opacus 1CP cells was determined by means of a biosensor method with the use of both a suspension of freshly harvested intact cells and immobilized resting cells [23, 24]. The advantages of the whole-cell biosensor method for determination of enzyme activity are small amounts of microbial biomass for biosensor system formation and rapidness of assay.
Besides the measurement of the enzyme activity, whole cell biosensors can be used for detection of toxic aromatic compounds. To improve selectivity of a microbial sensor, whole-cell bacterial bioreporter sensors for aromatic compounds are being developed. In a bioreporter sensor, a genetically encoded reporter protein is produced in response to a contact of a microbial cell with an analyte [25, 26, 27]. The main limiting factor for construction of whole-cell bacterial bioreporter sensors for chloroaromatic compound is shortage of knowledge about metabolic (biochemical) pathways of bacteria capable of degrading chloro-substituted aromatic compounds [26].
In this study, laboratory models of the membrane microbial sensor were formed on the basis of immobilized cells of R. opacus 1CP, which contained enzymes of degradation of benzoate or 3-chlorobenzoate (benzoate 1,2-dioxygenase or 3-chlorobenzoate 1,2-dioxygenase, respectively), to explore features of microbial metabolism of toxic aromatic compounds. The models were applied to evaluate the actinobacterial cells’ responses to benzoate and 3-chlorobenzoate in the absence of both enzymes, benzoate 1,2-dioxygenase and 3-chlorobenzoate 1,2-dioxygenase, or in the presence of one of the said enzymes.
The object of our study was a Gram-positive non-spore-forming R. opacus 1CP actinobacterium (DSM 46757, and VKM Ac-2638), which was isolated from the selective medium with 2,4-dichlorophenol. The culture was able to degrade benzoate (BA) and some of substituted benzoates [15]. The actinobacterium was maintained on agarized Luria-Bertani (LB) medium and transferred every 6 months.
The culture was grown on Petri dishes containing agarized LB medium (the medium
without 3CBA and BA) or agarized mineral medium with BA (200 mg/L) as a sole
carbon and energy source. The mineral medium had the following composition (g/L):
Na
Grown R. opacus 1CP cells were centrifuged (12,000–16,000 g, 10–15 min, + 4 °C) and washed twice with a 50-mM K-Na phosphate buffer (pH 7.4). BA-grown biomass and one part of LB-grown biomass were suspended in the phosphate buffer up to 100 mg of wet cells per mL, stored at + 4 °C for 12 h and were then used for preparation of receptor elements on the basis of BA- or LB-grown cells, respectively. Another part of LB-grown biomass was induced by BA or 3CBA.
To induce BDO or 3CBDO in LB-grown R. opacus 1 CP cells, LB-grown biomass was suspended in the phosphate buffer containing 2 g/L of BA (for BA-induced cells) or in the phosphate buffer containing 100 mg/L of 3CBA (for 3CBA-induced cells), respectively. The culture cells were incubated at 28 °C on a rotary shaker (n = 220 rpm) for 96 h (for BA-induced cells) or 24 h (for 3CBA-induced cells). After incubation, biomass (BA- or 3CBA-induced biomasses) was centrifuged and washed twice with the phosphate buffer. BA- and 3CBA-induced biomass were suspended in the phosphate buffer up to 100 mg of wet cells per mL, stored at + 4 °C for 12 h and were then used for preparation of receptor elements based on BA- or 3CBA-induced cells, respectively.
Bioreceptors were formed on the basis of obtained suspensions (100 mg of wet
cells weight per mL) of the actinobacterial cells. To form a bioreceptor,
bacterial suspension was immobilized on Whatman paper by the method of physical
adsorption. To achieve it, actinobacterial suspension (10
Cells’ responses to substrates were determined by biosensor method using
laboratory models of the membrane microbial sensor. Each laboratory model was
formed on the basis of the bioreceptor using R. opacus cells either LB-
or BA-grown, BA- or 3CBA-induced cells. To form a recognizing part of the
biosensor, the bioreceptor was fixed on the measuring surface of the Clark-type
oxygen electrode by means of nylon net. The oxygen electrode with the bioreceptor
(a microbial electrode) was placed in an open 5 mL measuring cell. The cell was
filled with an air-saturated buffer and equipped with a magnetic stirrer. The
design of the recognizing part of the membrane microbial sensor is presented in
our previously published paper [28]. The Clark-type oxygen electrode was used as
a transducer of the response of microbial cells. An amplifier system (Ingold
531-04 O
The measurements of cells’ responses to substrates were performed in the air-saturated phosphate buffer at + 20–22 °C at permanent stirring. The microbial electrode was placed in the measuring cell with the buffer solution and basal (endogenous) cell respiration was then stabilized. After stabilization of endogenous respiration of R. opacus cells, substrate solution was injected into the buffer solution. The cells’ response to substrate was proportional to a rate of cells’ respiration change. The change of cells’ respiration led to a change in oxygen concentration at the measuring area of the Clark-type oxygen electrode. The Clark-type oxygen electrode transduced a chemical signal (the change in oxygen concentration) into electric signal (a change in the electrode current). The change in the electrode current was recorded with the two-coordinate recorder. The recorded signal reflected the rate of change in the electrode current (dI/dt, pA/s), which was proportional to a change in oxygen consumption by culture in response to substrate injection. The cells’ response to substrate was calculated as the first derivative of the electrode current change in response to substrate addition (pA/s). After measurement, the system was washed, the basal respiration was then registered, and the system was ready to make the next measurement.
To measure the response to substrate for another R. opacus 1CP cells using another bioreceptor, an appropriate bioreceptor was fixed on the measuring surface of the Clark-type oxygen electrode, and measurements were made.
The presented data are the results of one of two/three independent experiments.
The measurements were taken in triplicate. Presented results are average values.
Statistical data analysis was carried out using a Student’s t-test
taking p
Our earlier study showed that BDO and 3CBDO of R. opacus 1CP are inducible enzymes, which are synthesized in the cells grown in the presence of substrates of enzymes [23, 29]. Furthermore, in our previous work, the response to the substrate for intact R. opacus 1CP cells used as a recognizing element of a reactor microbial sensor was an indicator of the activity of the enzyme initiating degradation of the substrate, BDO or 3CBDO [24]. LB medium is the BA- and 3CBA-free medium. Therefore, LB-grown actinobacterial cells contained only insignificant amounts of inducible enzymes (basal activity), which could not be registered in microbial cells.
It is known that the response to a substrate for immobilized cells of a bioreceptor of a membrane microbial sensor is caused by processes of both transport of a substrate into microbial cells and metabolism of a substrate in cells [30]. It is obvious that in the absence of the enzyme, which initiates substrate degradation, immobilized cells’ response to a substrate is caused by processes of substrate transport into cells. When no BDO and 3CBDO activity were registered in intact LB-grown R. opacus 1CP cells, immobilized cells’ response to BA or 3CBA was caused by transport of a substrate into LB-grown cells. Thus, curves shown in Fig. 2 describe a change in the rate of BA and 3CBA transport into R. opacus 1CP cells. Detection of the response to BA and 3CBA for 1CP actinobacterial cells non-induced by the substrate was the evidence of the presence of a constitutive system for substrates transport into cells. Earlier, both for Gram-negative bacteria and substrate other than BA, permeability of non-induced and induced cells to substrate was registered by Hegeman for Pseudomonas putida ATCC 12633/mandelate [31] as well as a constitutive system of uptake was observed by Migues et al. [32] for Alcaligenes denitrificans BRI 6011/2,4-dichlorobenzoic acid.
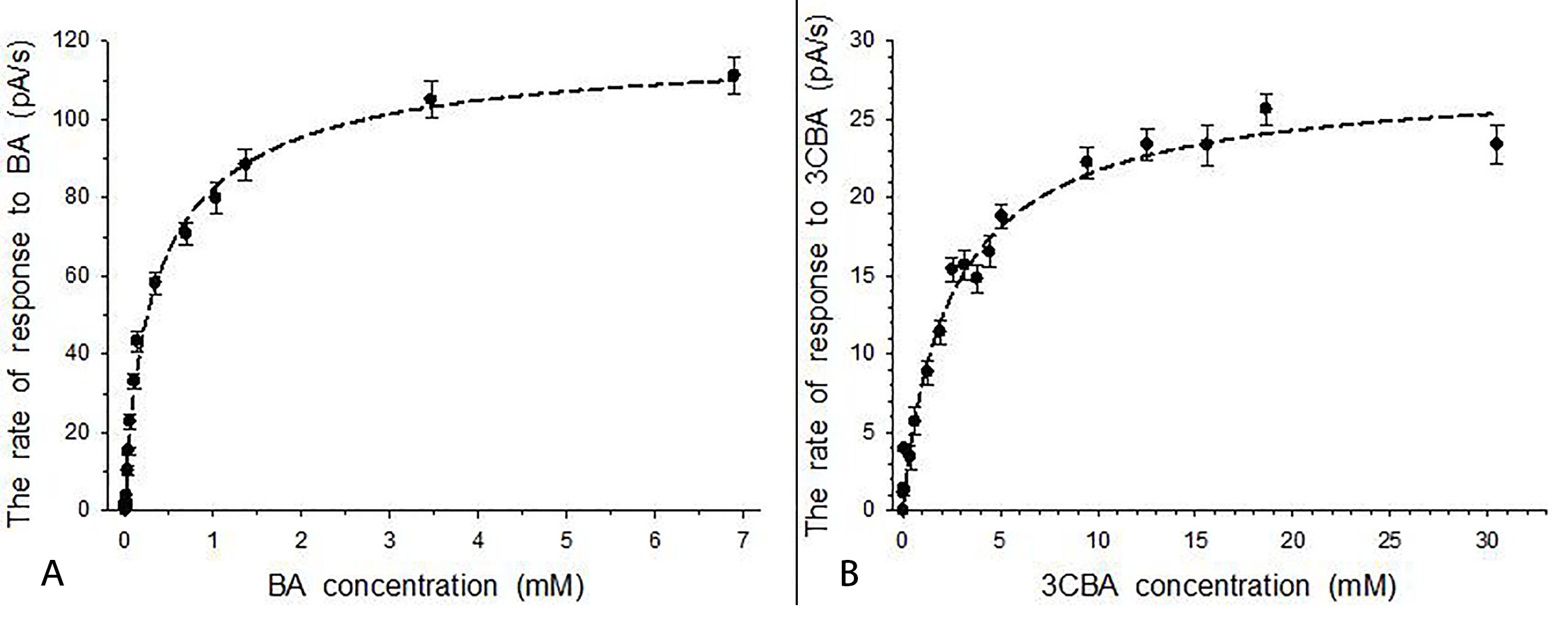 Fig. 2.
Fig. 2.Responses to the substrate for immobilized R. opacus 1CP cells grown in LB medium (BA- and 3CBA-free). (A) Responses to BA for BDO-free cells. (B) Responses to 3CBA for 3CBDO-free cells. Error bars indicate standard deviation from the measurement of three samples.
LB-grown cells of the culture are BDO- and 3CBDO-free cells. When BA and 3CBA penetrate the LB-grown cells, these substrates cannot be metabolized by the cells. However, these substrates were not accumulated in the cells. It was supported by the fact that receptors based on immobilized R. opacus 1CP cells, which was not induced by substrate, operated for longer than 24 h without loss of activity and sample throughput [33] of the membrane microbial sensor was 4–5 samples per hour. Hence, BA and 3CBA should have entered and exited LB-grown 1CP cells. As for substrate-induced cells of Pseudomonas putida ATCC 12633 in the presence of mandelate, concentrating permease activity was shown by Higgins and Mandelstam [34]. Since BA and 3CBA were not accumulated in the cells, constitutive concentrating permease could not mediate transport of these substrates into the cells of 1CP culture.
Aubert and Motais demonstrated that weak benzoic acid penetrates the cell
through the membrane lipids as undissociated acid [35]. In the present study,
cells’ responses were measured in the buffer at pH 7.4. In aqueous solution at pH
7.0, BA was dissociated by approximately 99.9% [36], and significant amounts of
BA (as anion) were unable to independently diffuse into the cell through simple
diffusion. Saturation kinetics (Fig. 2) was observed for dependency of the rates
of R. opacus 1CP response to BA or 3CBA on BA or 3CBA
concentration, respectively. It excluded the involvement of simple (passive)
diffusion in BA and 3CBA transport into LB-grown 1CP cells (linear regression
between the rate and concentration would be obtained). At the same time, active
transport of BA and 3CBA was hardly improbable for resting immobilized cells
knowing that the active transport is coupled with ATP hydrolysis. Most likely,
carriers-mediated diffusion facilitated BA and 3CBA transport into non-induced 1CP cells. The question remains unaswered whether porins like BenF
and BenP (porins can also be present in a wall zone of Gram-positive bacteria
[37]) and membrane BA transport proteins, such as BenE1 and BenE2 (2.A.46
benzoate:H
Nonlinear regression fit of data to Michaelis-Menten equation resulted in
calculation of rate constants V
Both cells induced with BA in non-growth conditions (referred as BA-induced cells) and cells grown in BA-medium (referred as BA-grown cells) contained BDO initiating BA metabolism in cells. Therefore, the response to substrate for BDO-containing immobilized R. opacus 1CP cells was caused by both the process of substrate transport into cells and process of initial metabolism of substrate in cells. Due to specificity of BDO of 1CP actinobacterium, responses to both substrates (BA and 3CBA), which characterized BDO activity, were registered earlier using intact cells [10]. In the present study, responses to BA (Fig. 3A and 3B) and 3CBA (Fig. 3C and 3D) were studied using BDO-containing immobilized cells of the culture. The cells’ response to BA was markedly higher than the cells’ response to 3CBA. It could be a result of a drop of enzyme activity (BDO activity) in the presence of 3CBA as a substrate. Similar change of enzyme activity was shown in previous research with intact cells: BDO activity in the presence of BA was higher by a factor of 5 than in the presence of 3CBA [10]. It is a similar situation as described by Krooneman et al. [42] for determination of maximum oxygen uptake rate in the presence of BA or 3CBA for BA-grown Alcaligenes sp. L6. For this culture in the presence of BA, cells’ response was a 5-fold higher than a response in the presence of 3CBA. The difference between cells’ responses to BA and 3CBA can also be explained by the presence of different transporters in 1CP cells in the presence of BA or CBA. For instance, various transporters were induced by BA (BenK, BenE1 and BenE2) and 3CBA (BenK and BenE2) in Pseudomonas putidaKT2440 cells [43]. Furthermore, in confirmation of the above said about the response to substrate for immobilized 1CP cells, the categories of expressed genes such as ‘energy metabolism’ and ‘transport and binding proteins’ characterized the cellular responses to substrate for Pseudomonas putida KT2440 cells reported by Wang et al. [43]. In addition, Clark et al. and Wong et al. [40, 44] showed that transport process and process of substrate metabolism are dependent of each other.
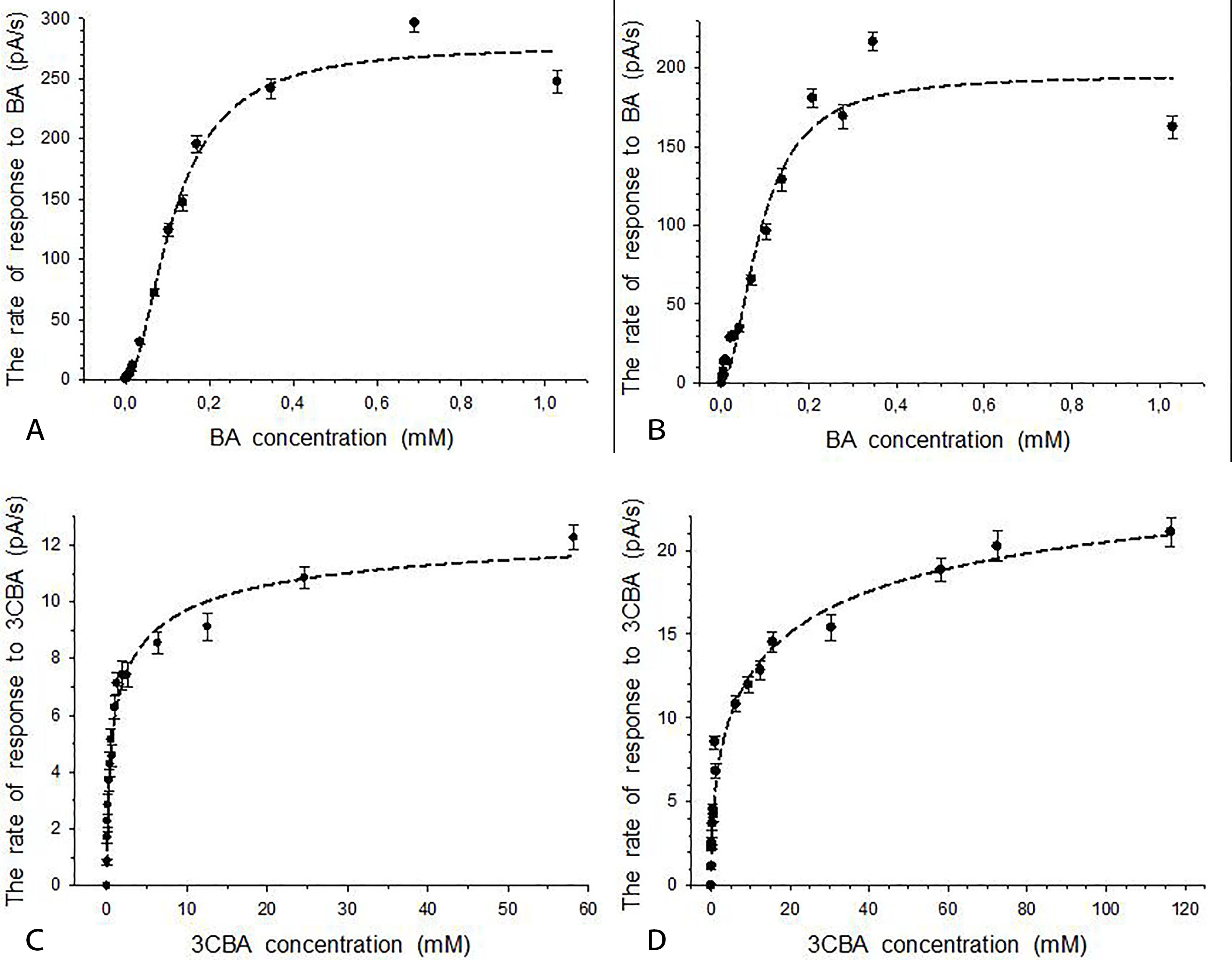 Fig. 3.
Fig. 3.Responses to BA or 3CBA for immobilized R. opacus 1CP cells in the presence of BDO in cells. (A) Responses to BA for BA-induced cells. (B) Responses to BA for BA-grown cells. (C) Responses to 3CBA for BA-induced cells. (D) Responses to 3CBA for BA-grown cells. Error bars indicate standard deviation from the measurement of three samples.
Curves of dependencies of the cells’ response to substrate (both BA and 3CBA) on
substrate concentration for BA-induced and BA-grown immobilized cells are
represented in Fig. 3. Saturation curves were obtained. Nonlinear regression fit
of data to the Hill equation was found for all curves. Half-saturation constants,
S
Regarding the Hill kinetics for cells’ responses to BA, positive kinetic cooperativity by the substrate was found for responses of BA-induced and BA-grown immobilized cells. Values of Hill coefficient (n) were greater than 1. They were 1.87 and 1.90 for BA-induced and BA-grown 1CP cells, respectively. It was in conformity with dependency of cooperatives by the substrate for BDO on BA concentration in the growth medium for R. opacus 1CP cells. This phenomenon was found in our study using intact cells [45]. For BDO of intact 1CP cells, positive kinetic cooperativity by BA was detected when BA concentration in the growth medium was lower than 6 g/L. Furthermore, considering the dependency between substrate transport process and process of substrate metabolism [44], positive kinetic cooperativity was observed by Choudhary et al. [38] for BA transport into Pseudomonas putida CSV86 cells (n = 1.9).
For cells’ responses to 3CBA in the presence of BDO, unlike cells’ responses to BA, negative kinetic cooperativities by the substrate were detected. n values were 0.54 and 0.51 for BA-induced and BA-grown 1CP cells, respectively. Responses to 3CBA were measured for the same cells induced by BA, which were used for detection of responses to BA. In both experiments (both with BA and with 3CBA) the same enzyme (BDO) caused the cells’ response to substrate. Therefore, allosteric interaction of active sites of BDO in the presence of 3CBA could be a reason of negative kinetic cooperativities by substrate [46].
Another reason of negative kinetic cooperativity could be the presence of two phases (hyperbolic and linear) for ‘response-3CBA concentration’ dependency that was obtained for BA-induced cells (Fig. 4). Linear phase indicated that the simple diffusion took part in formation of the cells’ response to 3CBA when 3CBA concentration was higher than 10 mM.
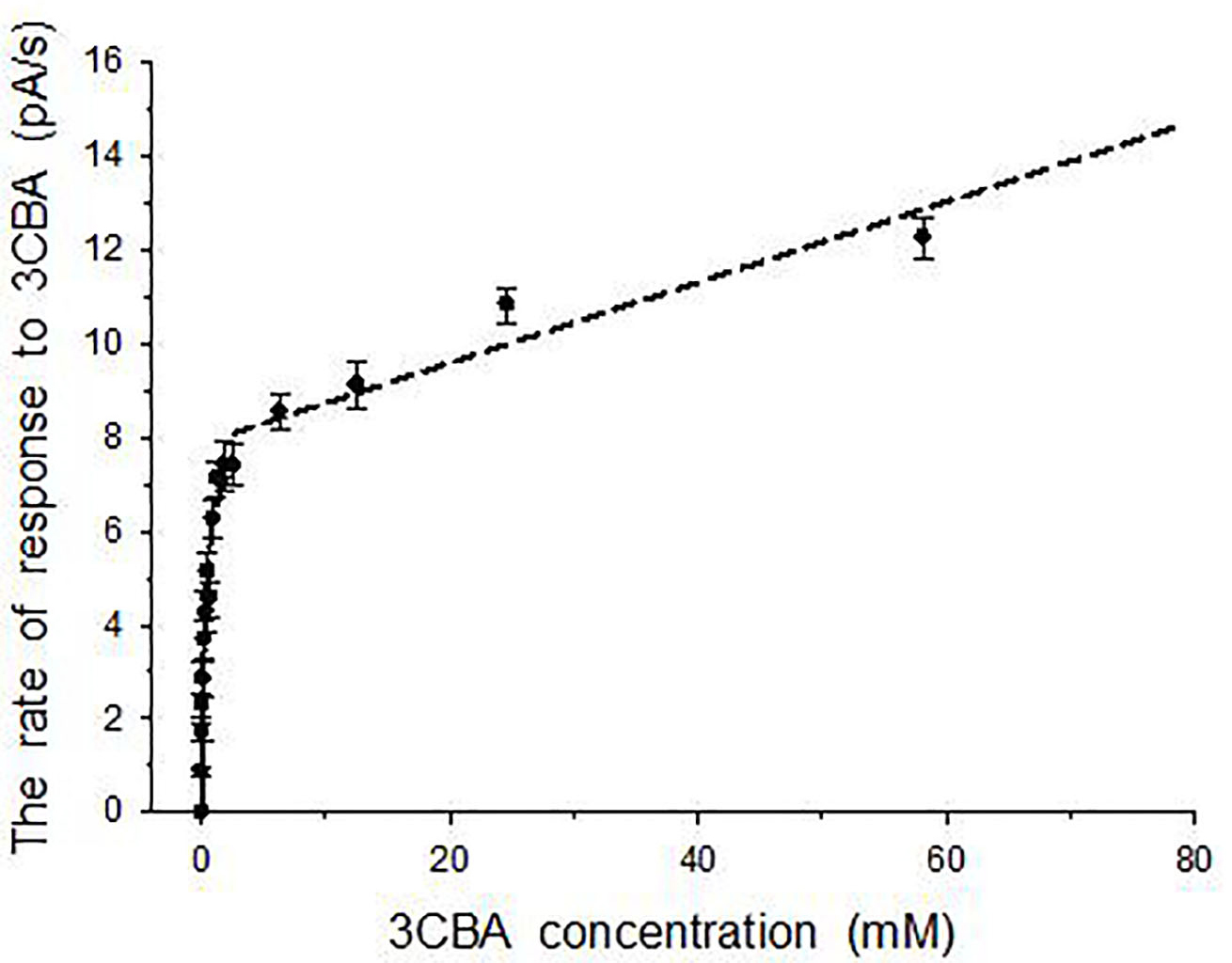 Fig. 4.
Fig. 4.Two phases dependency of the rate of the cells’ response to 3CBA on 3CBA concentration for BA-induced R. opacus 1CP cells. Error bars indicate standard deviation from the measurement of three samples.
After cells induction in the presence of 3CBA, R. opacus 1CP cells contained 3CBDO. For these immobilized cells, the response to 3CBA was caused by 3CBA transport and 3CBDO activity. A curve of the dependency of the cells’ response to 3CBA on concentration of 3CBA is shown in Fig. 5.
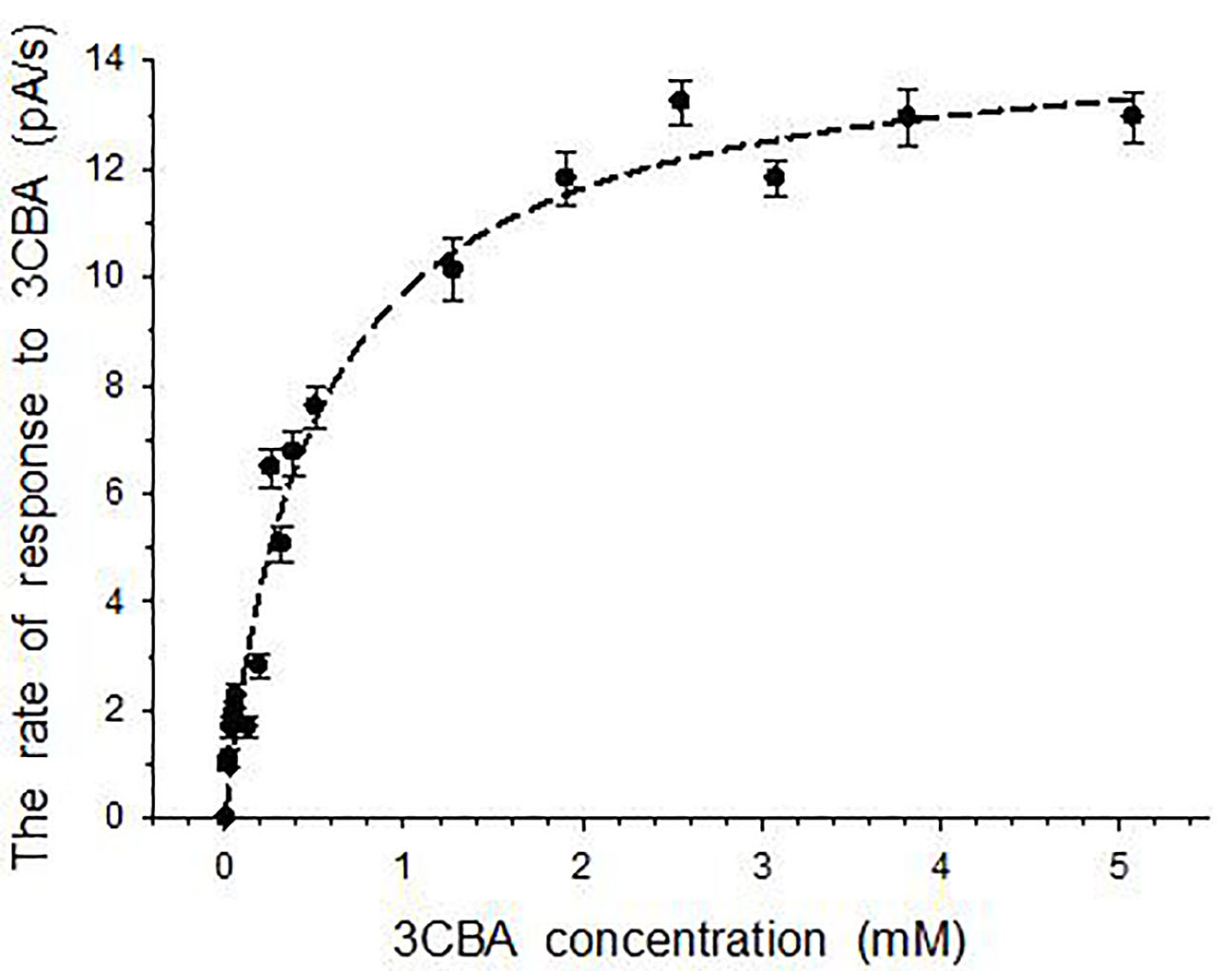 Fig. 5.
Fig. 5.Responses to 3CBA for immobilized R. opacus 1CP cells induced by 3CBA. Error bars indicate standard deviation from the measurement of three samples.
Saturation curve was obtained; data of nonlinear regression fitted to the
Michaelis-Menten equation. Half-saturation constants (S
In sum, laboratory models of the membrane microbial sensor, based on immobilized Rhodococcus opacus 1CP cells and the Clark-type oxygen electrode as a transducer, were successfully applied to evaluate the actinobacterial cells’ responses to BA and 3CBA.
The data from this study show that the membrane microbial sensor based on immobilized cells of the bacterial culture could be useful as a perspective tool for evaluation of enzymes of complex structure initiating substrate metabolism coupled with oxygen consumption.
The Rhodococcus opacus 1CP culture has constitutive system(s) for the transport of BA and 3CBA into cells. Therefore, cells without prior induction can be introduced into an environment contaminated with these xenobiotics. The positive cooperativity by the substrate, which was shown for immobilized cells of the culture, indicates that the catalytic efficiency of the active sites of the enzyme (BDO) increases as they are filled with the substrate. Moreover, adaptation to one substrate (BA or 3CBA) does not preclude the use of the second substrate. In the light of it, we can conclude that Rhodococcus opacus1CP is an ideal xenobiotics-degrading culture for remediation of soil contaminated by BA and 3CBA.
The results of the study have indicated a direction for our further research. Previously, the presence of porins was reported only for Gram-negative bacteria. Currently, pore-forming proteins have been found in Gram-positive bacteria, including rhodococci. There is no data on the pore-forming proteins of Rhodococcus opacus 1CP. Microbiological studies are planned to confirm or refute the presence of pore-forming proteins in the wall zone of Gram-positive Rhodococcus opacus 1CP. In addition, genetic studies will help identify genes encoding BA and 3CBA transporters.
BA, benzoate; BDO, benzoate 1;2-dioxygenase; 3CBA, 3-chlorobenzoate; 3CBDO, 3-chlorobenzoate 1;2-dioxygenase.
EVE and IPS conceived and designed the experiments, and performed the experiments; EVE analyzed the data; EVE and IPS contributed reagents and materials. Both authors read and approved the final manuscript.
Not applicable.
Thanks to all the peer reviewers for their opinions and suggestions.
This research received no external funding.
The authors declare no conflict of interest.
