Academic Editor: Yiannis Kourkoutas
Background: The high salt concentration is the major factor limiting microbial growth at salterns, along with solar radiation, temperature, and pH. These environmental factors play key roles in the acquisition of unique genetic adaptations for the survival of microorganisms in salterns, which can result in the production of interesting secondary metabolites. The main goal of the present work was to isolate and compare the culturable microbiota from two geographically distant salterns in Portugal and access their biotechnological potential. Methods: Culturomics approaches using different culture media were applied for microbial isolation. All isolates were identified either by 16S rRNA or ITS genes sequencing, and their biotechonological potential was assessed by PCR. Results: Overall, 154 microbial isolates were recovered that were phylogenetically assigned to 45 taxa from 9 different phyla. From these, 26 isolates may represent putative new taxa. The predominant genera obtained were Penicillium (41 isolates, 26.6%), Streptomyces (13 isolates, 8.4%) and Sinomicrobium (11 isolates, 7.1%). Moreover, the polyketide synthase I gene was present in 64 isolates, the nonribosomal peptide synthethase gene in 16 isolates, and both genes in 23 isolates. Conclusions: This study adds up valuable knowledge on the culturable microbiota of Portuguese salterns and on its potential for production of secondary metabolites. In the long run, this study provides a widely diverse microbial collection for future works. Data public repository: All DNA sequences were deposited in the GenBank database at National Centre for Biotechnology Information (NCBI) web platform under accession numbers OK169439-OK169485, OK216020-OK216124, OK287059 and OK326927.
Portugal has an extensive coastline harbouring a wide range of different environments, including solar salterns. In Portugal, the production of salt in traditional salterns, especially in the North, takes place in the summer season. During salt production, saltern waters are hypersaline environments characterized by high concentrations of NaCl, UV radiation, temperature and pH. These factors are determinant in biodiversity modulation [1], narrowing the microbial community to well adapted halophilic (extremophiles) or halotolerant microorganisms. The microbial diversity of hypersaline environments has been targeted by the scientific community along the last decade [1, 2, 3, 4, 5, 6, 7]. The abundance of microorganisms from different phyla was acknowledged on this type of extreme environment, comprising members of Euryarchaeota, Planctomycetota, Bacteroidota, Rhodothermota (previously included in Bacteroidota [8]), Pseudomonadota, Actinomycetota and Cyanobacteria [2, 3], Bacillota [7, 9], Gemmatimonadota [6] and Eukaryota as microalgae [10] and Ascomycota [11, 12].
The ability of microorganisms to produce natural products (NPs) with relevant biotechnological value is well recognised [13] and NPs obtained from extremophiles have proved their biotechnological value in a wide range of fields [14]. As extremophiles are often exposed to sudden and repeated fluctuations derived from global changes, such as temperature and water availability, they require a great physiological adaptability at different cellular levels, namely in their biological membranes, proteins and extracellular metabolites [14]. Due to these metabolic adaptations, the unexploited microbiota of salterns presents a high potential for novel NP discovery with applications in important biotechnological fields, such as medicine, pharmaceutics, cosmetics, agriculture, and the food industry [15, 16, 17, 18].
One of the currently applied molecular methodologies to assess the potential of microorganisms to produce bioactive molecules consists in Polymerase Chain Reaction (PCR) protocols. Genes of nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) are commonly targeted as a preliminary screen of bioactive properties, since they are well known to be responsible for the production of enzymatic complexes that are involved in the synthesis of NPs. Specifically, NRPSs are extensively associated with the production of structurally diverse peptides presenting a wide range of activities, like antibacterial, antitumour, cytostatic or immunosuppressive [19], while PKSs are engaged in the assembly of distinct NPs – the polyketides. This class of NPs includes many biotechnologically important compounds such as antibiotics, antiparasitics, anticancer, immunomodulators, antifungals and anticholesteremics [20]. In fact, the annual sales of medicines derived from polyketides had already reached 20 billion US dollars [21].
The microbial diversity of Portuguese salterns has been poorly studied. Few
studies were obtained by culture independent approaches [22, 23], and a few more
through culture dependent methods [24, 25, 26]. The present study targeted the
culturable microbial diversity, in late summer of 2018, of two Portuguese
salterns, Aveiro and Olhão salterns. These have different geographical
locations: Aveiro is situated in the North, being influenced by the Atlantic
Ocean, while the Olhão saltern is in the South, and is exposed to the
influence of not only the Atlantic Ocean but also the Mediterranean Sea. Average
temperature and pluviosity levels per year are also different at both locations:
15.6
Fresh and wet salt, sediment from under the salt and water, all sampled directly
from salt production ponds, were collected from Aveiro saltern
(40º38’50” N, 8º39’46” W) in September 2018
and from Olhão (37
 Fig. 1.
Fig. 1.Representation of the sampling locations along with the overall culturomics methodologies applied.
Media M600, M607 [28] and M607SW (this study) (Supplementary Table 1),
supplemented with 200
Salt samples were dissolved in autoclave-sterilized seawater (SW) until
saturation and serially diluted until 10
Concerning sediment samples, 1 g of each saltern sediment was individually
suspended in 1 mL of sterile SW by vigorously vortexing. From this suspension,
serial dilutions until 10
Agar media were used for direct isolation, while broth versions of the media
were used for enrichments. Specifically, enrichments were carried out in 24-well
plates containing 900
Agar media regularly used for Actinomycetota isolation were selected, namely R2A-Agar (BD Difco™, Maryland, USA), Starch-Casein-Nitrate agar (SCN; adapted from Küster and Williams [29]), M3 [30] and a modified Nutrient-Poor Sediment extract agar (NPS; adapted from Jensen et al. [31]) (Supplementary Table 1). All agar media were supplemented with 50 mg/L cycloheximide (Batch: 9I011709; Sigma-Aldrich, Missouri, United States), 50 mg/L nalidixic acid (Batch: 8D012744; AppliChem) and 50 mg/L nystatin (Batch: 9J012880; Sigma-Aldrich).
Salt samples were dissolved in 5 mL of sterile SW until saturation, while 1 g of
each sediment sample was suspended in 2 mL of sterile SW by shaking at 200 rpm
for 30 min and by vortexing at maximum speed for 5 min. Two mL of the salt brown
residual liquid and of the water samples were directly used as inoculum. All
these samples were exposed to a heat pre-treatment at 60
Incubation of samples with calcium carbonate (CaCO
Potato Dextrose Agar (PDA; BD Difco™) is widely used for Fungi cultivation and was the medium selected for the present study, which was prepared with SW. All samples, namely salt, salt brown residual liquid, sediment and water, as well as the serial dilutions of these samples were prepared as described above for isolation of Planctomycetota. All these suspensions, either diluted or not, were individually plated in PDA.
In all methodologies previously described, bacterial colonies with different
morphologies were restreaked until the achievement of axenic cultures. All pure
isolates were cryopreserved in 24% glycerol at –80
Each axenic isolate was cultivated in the corresponding broth medium of
isolation and these cultures were used for DNA extraction. For isolates obtained
in PDA medium, NZY Plant/Fungi gDNA Isolation kit (NZYTech, Lisbon, Portugal) was
used, according to the manufacturer’s instructions. In the case of the isolates
obtained in the remaining media, DNA was extracted using the E.Z.N.A. Bacterial
DNA kit (Omega Bio-Tek, Norcross, Georgia, USA), according to the manufacturer’s
instructions. DNA quantification was done by using the
Bacterial DNA was PCR-amplified with the universal primers 27F and 1492R [36], while Fungi DNA was PCR-amplified by using ITS1 and ITS4 primers [37].
For the isolates which strain designation starts with “C.”, the 16S rRNA gene
primers, 27F and 1492R, were used and the PCR mixture consisted of 1
For the remaining isolates, including Fungi, the PCR mixture (25
Sequence analyses were carried out using Geneious Prime (Biomatters Ltd, Auckland, New Zealand). Curated sequences were matched in the EzBioCloud [38] database to determine their closest relatives. Phylogenetic trees were built using MEGA 7 (Pennsylvania State University, Pennsylvania, USA) software [39]. Briefly, all sequences, either from the isolates in this study and from the closest related strains determined on EzBioCloud, were grouped by phyla. Separately, phyla-grouped sequences were aligned using the ClustalW algorithm with a Gap Open Penality of 15.00 and a Gap Extension Penalty of 6.66. The resulting multiple alignments for each phylum were used to construct phylogenetic trees, by applying the Maximum Likelihood statistical method, the phylogeny test based on the Bootstrap method considering 1000 replicates, and the Tamura-Nei substitution model. Different strains were used as outgroup depending on the phyla.
All sequences obtained on this study were submitted to the GenBank database [40] at National Centre for Biotechnology Information (NCBI) web platform under the following accession numbers: OK169439-OK169485, OK216020-OK216124, OK287059 and OK326927.
PAST 3.22 (University of Oslo, Oslo, Norway) [41] software was used to calculate the Fisher’s
A sample rarefaction curve was estimated in PAST 3.22 by using the Mao’s Tau index [42]. Similarities among samples were calculated on PAST 3.22 by using the Sørensen coefficient and Bray-Curtis index. All results were computed taking into account a 95% confidence and 1000 iterations as bootstrap value.
The putative potential of all isolates obtained in this study to produce NPs was
assessed through PCR. Briefly, isolates DNA was screened for the presence of NRPS
and PKS-I genes. NRPS genes were amplified using primers MTF2 [5
Aiming at a broader range of microbial isolation, 6 media with different nutritional compositions along with 2 pre-formulated media (Supplementary Table 1) were assayed. A medium selective for fungi, PDA, allowed the isolation of 45 fungal strains. The most effective media for bacteria were M600 (32 strains), R2A (32 strains) and M607 (31 strains). The remaining media used were less rich in nutrients and apparently less successful (14 strains altogether in 4 different media). Ultimately, the media used proved to be effective in the isolation of a highly heterogenous microbiota from salterns.
The microbial isolation screening carried out for samples from Aveiro and Olhão salterns allowed the isolation of 154 strains, of which 47 were affiliated with Fungi and 107 with Bacteria. Particularly, 26 fungal and 75 bacterial isolates were obtained from Aveiro, and 21 fungal and 32 bacterial isolates from Olhão. All these isolates have been putatively identified based on the ITS and 16S rRNA genes for Fungi and Bacteria, respectively. The isolates relatedness was examined by building phylogenetic trees (Supplementary Fig. 1).
Despite the high abundance of salterns in Portugal, their microbiota has been poorly explored. The existent research reports comprises (i) the description of new strains detected by metagenomics approaches of Tavira saltern (Algarve salt flats) microbiota [46], (ii) metagenomics studies of Algarve salt flats [22, 23, 47], (iii) characterization of novel isolated species from Tavira saltern [26] and from Tejo salt flats [48], (iv) an archaeal isolate genome announcement from Figueira da Foz salt flats [49] and (v) extensive isolation approaches targeting Aveiro saltern and a saltern from Olhão (Algarve salt flats) focusing on the overall bacterial microbiota [24]. In this last study, the direct comparison between the microbiota of both salterns was not possible since the nature of samples, the methodology applied and the time of sampling were different between both salterns. None of these studies have targeted the fungal community, most likely because the prevalence of fungi in marine and aquatic environments, including hypersaline ones, was overlooked for many years [50, 51, 52]. In the last decades, Fungi members have been reported in every aquatic environment explored, including marine and hypersaline ones [50, 51, 52]. The first description of fungi in salterns was reported by Gunde-Cimerman and collaborators [53], which aroused a fascinating interest in the scientific community [12, 54, 55]. Since this, saltern fungi have been studied worldwide, however the Portuguese salterns mycobiota remains unknown. In this study, the microbial diversity was assessed at genera (Fig. 2) and phyla (Fig. 3) levels. The fungal isolates obtained were phylogenetically related with the phylum Ascomycota (Fig. 3), with the exception of isolate AW17 that was affiliated to the genus Sporobolomyces (Fig. 2), specifically to the species Sporobolomyces ruberrimus, a yeast within the phylum Basidiomycota (Fig. 3).
 Fig. 2.
Fig. 2.Microbial diversity of the isolates obtained from Aveiro and Olhão salterns represented by genera.
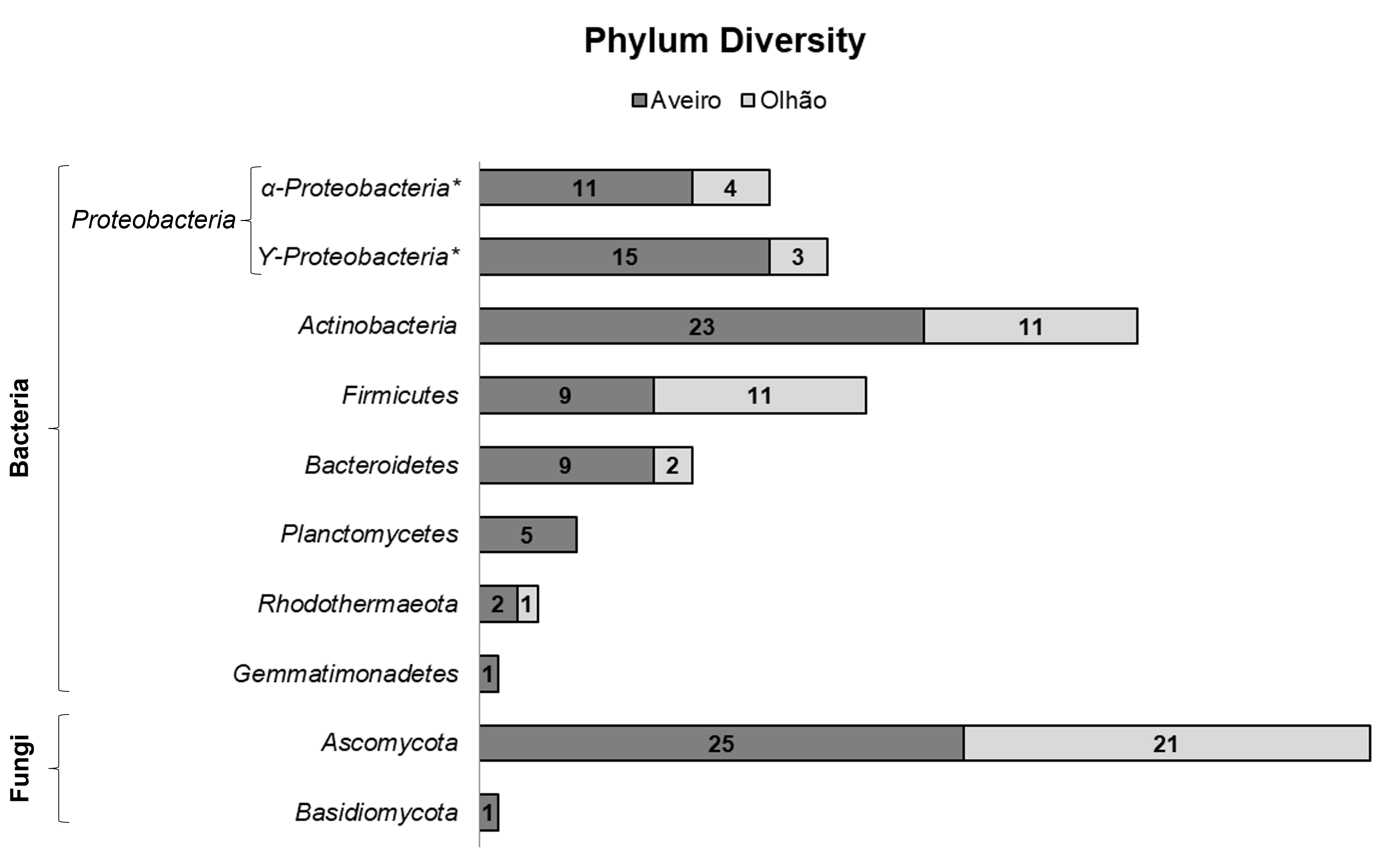 Fig. 3.
Fig. 3.Overall microbial biodiversity, phyla-ranked, of the isolates obtained from Aveiro and Olhão salterns, belonging to two microbial kingdoms.
Colour scheme representation by phylum: grey for Ascomycota, black for Basidiomycota, blue for Pseudomonadota, orange for Actinomycetota, purple for Bacillota, brown for Bacteroidota, green for Planctomycetota, yellow for Rhodothermota and red for Gemmatimonadota. Full filled with colour represent isolates common to both salterns and filled with pattern represents exclusivity to a specific saltern. (*) represents isolates identified only to the family level.
Overall information of the environmental prevalence of strains phylogenetically closely related with the isolates obtained with the present study is compiled in Table 1 (Ref. [1, 5, 9, 11, 24, 26, 53, 54, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107]), where presence or absence in salterns environments was highlighted.
| Kingdom | Phylum | Class | Order | Family | Closest related microorganism | Isolates from this study | Environments reported |
| Fungi | Ascomycota (Supplementary Fig. 1a) | Dothideomycetes | Pleosporales | Pleosporaceae | Alternaria alternata | ASBR4 | Salterns [56] |
| Dothideales | Dothioraceae | Aureobasidium pullulans | ASED7 | Salterns [53, 57, 58] | |||
| Capnodiales | Davidiellaceae | Cladosporium herbarum | AW2 | Salterns [54, 59] | |||
| Cladosporium perangustum | AS110 | Marine [60] | |||||
| Cladosporium phaenocomae | AS102 | Plants [61], deteriorated wood [62] | |||||
| Eurotiomycetes | Eurotiales | Trichocomaceae | Penicillium brevicompactum | ASED17, ASED27, ASED28, ASBR9, AS103, AS104, AS108, AS111, OSBR109, OS1, OS3, OS4, OS6, OS7, OS8, OW12 | Salterns [54] | ||
| Penicillium brocae | ASED21, ASED25, ASED29, OSBR107, OSBR117, OS2, OS5, OS10, OW1, OW2, OW3, OW8, OW17 | Marine [63] | |||||
| Penicillium chrysogenum | ASED16, ASED18, ASED19, ASBR1, ASBR2, ASBR5, AW1, AW11, OSBR108, OW5 | Salterns [11, 54, 64] | |||||
| Penicillium dierckxii | ASED8 | Hypersaline [65] | |||||
| Penicillium sp. | OSBR118 | Plants (NCBI: EF694632) | |||||
| Basidiomycota (Supplementary Fig. 1b) | Microbotryomycetes | Sporidiobolales | Incertae sedis | Sporobolomyces ruberrimus | AW17 | Psychrophilic [66, 67, 68] | |
| Bacteria | Actinomycetota (Supplementary Fig. 1c) | Actinobacteria | Micrococcales | Dermabacteraceae | Brachybacterium paraconglomeratum | C.OS8 | Hypersaline [69, 70] |
| Brevibacteriaceae | Brevibacterium sediminis | C.AS1, C.AS9 | Salterns [24] | ||||
| Microbacteriaceae | Microbacterium aerolatum | C.ASBR1, C.OS5 | Air [71] | ||||
| Microbacterium amylolyticum | C.AS10, C.AW5 | Salterns [24] | |||||
| Microbacterium ginsengiterrae | C.AW1 | Soil [72] | |||||
| Micrococcaceae | Micrococcus luteus | C.OS4 | Salterns [73] | ||||
| Rothia kristinae | OSBR105, OSBR106 | Human commensal and opportunistic pathogen [74] | |||||
| Mycobacteriales | Gordoniaceae | Gordonia oryzae | AW6 | Plants [75] | |||
| Tsukamurellaceae | Tsukamurella tyrosinosolvens | OW4 | Human commensal and opportunistic pathogen [76] | ||||
| Propionibacteriales | Nocardioidaceae | Nocardioides salarius | OSBR100 | Marine [77] | |||
| Streptomycetales | Streptomycetaceae | Streptomyces intermedius | C.AS7, C.ASBR4, | Members of this genus have been associated with Salterns [9, 78, 79, 80] | |||
| C.AW3 | |||||||
| Streptomyces rubrogriseus | C.AS5, C.ASBR6, C.ASBR8, C.ASBR9, C.ASBR10, C.ASBR11, | Members of this genus have been associated with Salterns [9, 78, 79, 80] | |||||
| C.ASED8, C.OS6, | |||||||
| C.OS7, C.OSBR1 | |||||||
| Streptosporangiales | Nocardiopsaceae | Nocardiopsis lucentensis | C.ASBR7, C.ASED1, | Salterns [79, 81] | |||
| C.ASED2, C.ASED3 | |||||||
| Nocardiopsis prasina | C.ASED4, C.AW6 | Members of this genus have been associated with Salterns [9, 78, 79, 80] | |||||
| Acidimicrobiia | Acidimicrobiales | Iamiaceae | Uncultured bacterium | OSBR104 | Air [82] | ||
| Bacteroidota (Supplementary Fig. 1d) | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Psychroflexus tropicus | AW7 | Hypersaline [83] | |
| Sinomicrobium oceani | C.AS2, C.AS3, C.AS4, | Members of this genus have been associated with Salterns [1] | |||||
| C.AS6, C.AS8, C.ASED5, C.ASED7, C.ASED9, C.OS3, C.OS9 | |||||||
| Kingdom | Phylum | Class | Order | Family | Closest related microorganism | Isolates from this study | Environments reported |
| Bacteria | Bacillota (Supplementary Fig. 1e) | Bacilli | Bacillales | Bacillaceae | Alkalihalobacillus hwajinpoensis | ASED10, ASED13 | Salterns [24, 73, 84] |
| Bacillus safensis | AW4 | Salterns [85] | |||||
| Bacillus sinesaloumensis | AW3, AW5 | Human commensal [86] | |||||
| Cytobacillus luteolus | OSBR113 | Salterns [24, 84] | |||||
| Mesobacillus sp. | AW16, OSBR115 | Salterns [24] | |||||
| Metabacillus litoralis | OSBR101, OSBR112, OSBR114, OSBR116, OW6 | Salterns [84] | |||||
| Rossellomorea sp. | AW18, OW14 | Salterns [24] | |||||
| Thalassobacillus cyri | OSBR110, OSBR111 | Hypersaline [87]; Members of this genus have been associated with Salterns [73] | |||||
| Paenibacillaceae | Paenibacillus pabuli | ASED15, ASED30 | Members of this genus have been associated with Salterns [84] | ||||
| Planococcaceae | Paenisporosarcina quisquiliarum | OS9 | Soil [88]; Members of this genus have been associated with Salterns [84] | ||||
| Gemmatimonadota (Supplementary Fig. 1f) | Longimicrobia | Uncultured | Uncultured | Uncultured bacterium | AW12 | Members of this phylum have been associated with Salterns [73, 89] | |
| Planctomycetota (Supplementary Fig. 1g) | Planctomycetia | Planctomycetales | Planctomycetaceae | Alienimonas californiensis | ASED1 | Algae [90] | |
| Maioricimonas rarisocia | ASED14 | Marine [91] | |||||
| Pirellulales | Pirellulaceae | Rhodopirellula pilleata | ASED26 | Marine [92] | |||
| Rhodopirellula rubra | ASBR8 | Algae [93] | |||||
| Phycisphaerae | Phycisphaerales | Phycisphaeraceae | Uncultured bacterium | ASED31 | Members of this class have been associated with Salterns [26] | ||
| Pseudomonadota (Supplementary Fig. 1h) | Alphaproteobacteria | Hyphomicrobiales | Aurantimonadaceae | Aureimonas glaciistagni | OW13 | Members of this genus have been associated with Salterns [84] | |
| Caulobacterales | Caulobacteraceae | Brevundimonas sp. | ASED5 | Members of this genus have been associated with Salterns [94] | |||
| Rhodospirillales | Rhodovibrionaceae | Rhodovibrio sodomensis | OW16, OW18, ASBR14, ASBR15, ASBR16, AS100, AS106, AS114, AW21, AW23, OW9 | Salterns [95] | |||
| Rhodobacterales | Rhodobacteraceae | Roseibacterium elongatum | AW9 | Hypersaline [5] | |||
| Hyphomicrobiales | Stappiaceae | Stappia stellulata | ASED24 | Marine [96, 97] | |||
| Gammaproteobacteria | Oceanospirillales | Alcanivoracaceae | Alcanivorax dieselolei | C.AW4 | Members of this genus have been associated with Salterns [24] | ||
| Alcanivorax sp. | C.ASED6 | Members of this genus have been associated with Salterns [24] | |||||
| Alteromonadales | Alteromonadaceae | Marinobacter confluentis | AW10 | Marine [98] | |||
| Marinobacter sp. | ASED12 | Marine [99] | |||||
| Oceanospirillales | Halomonadaceae | Halomonas fontilapidosi | AW8, AW13 | Salterns [24, 100, 101] | |||
| Halomonas ventosae | ASED11 | Salterns [100, 101] | |||||
| Salinicola zeshunii | OW15 | Members of this genus have been associated with Salterns [102, 103, 104] | |||||
| Lysobacterales | Lysobacteraceae | Luteimonas padinae | ASBR3, ASBR7, ASBR10 | Salterns [105] | |||
| Cellvibrionales | Microbulbiferaceae | Microbulbifer halophilus | C.ASBR5, C.OW1, C.OW2 | Members of this genus have been associated with Salterns [106] | |||
| Chromatiales | Aquichromatiaceae | Uncultured bacterium | ASBR12, AW14, AW19, AW20 | Members of this genus have been associated with marine habitat [107] | |||
| Rhodothermota (Supplementary Fig. 1i) | Balneolia | Balneolales | Balneolaceae | Uncultured bacterium | ASBR13, AS101, OSBR103 | Members of this family have been associated with Salterns [1, 24] | |
| Those associated with saltern environment are highlighted in bold. | |||||||
Within Fungi, members of genus Penicillium were the only ones detected in both salterns and the most abundant ones (Figs. 2 and 3; Supplementary Table 2).
The Pseudomonadota genera Rhodovibrio and Microbulbifer were
the only ones common to both saltern (Fig. 2). The similarity score of all 4
Microbulbifer isolates with M. halophilus (
 Fig. 4.
Fig. 4.Phylogenetic framing of the putative novel taxa isolated from the two Portuguese Salterns. Maximum-Likelihood (ML) phylogenetic trees constructed with MEGA 7 software [39], using 16S rRNA gene sequences of the saltern isolates and of the phylogenetically closest sequences obtained in the EzBioCloud webserver. Bootstrap values were calculated based on 1000 replications and the numbers at each branch represent the bootstrap support in percentage for each cluster. The tree was constructed using representative members of each class within the phyla detected; Bacteroides fragilis (AB050106) was used as outgroup; Bar, 0.050 substitutions per nucleotide position.
Within the phylum Bacillota, Mesobacillus and Rossellomorea were the only two Bacillota genera that were detect at both Aveiro and Olhão salterns. Despite the geographical distance, the 2 strains (AW18 and OW14) closely affiliated with the novel established genus Rossellomorea [110] showed a perfect similarity between their 16S rRNA gene sequences and a close phylogenetic relationship with the isolate Rossellomorea sp. es.034 (PDIY01000001). Therefore, these strains are strong candidates for the description of a novel species (Fig. 4). Moreover, 2 isolates (AW3 and AW5) showed a similarity of up to 98.66% in the 16S rRNA gene with Bacillus sinesaloumensis, and other 2 isolates (OSBR114 and OW16) showed similarity scores of up to 98.4% in the 16S rRNA gene with Metabacillus litoralis. All of them may be representatives of novel taxa (Fig. 4).
Within Actinomycetota, 3 isolates were phylogenetically closely related with known opportunistic human pathogens (Rothia and Tsukamurella), that were obtained from Olhão saltern [74]. The presence of these species in Olhão saltern microbiota may be related with human activity, not only through salt exploitation but also through touristic related activities happening in Olhão salterns, like saltern baths (Table 1). Additionally, one isolate affiliated with Gordonia oryzae was obtained. This specie was recently described [75] and was not associated with extreme environments until this study. Furthermore, the Olhão saltern isolate OSBR104 was affiliated with the Actinomycetota family Iamiaceae, but the closest related strain described was Aquihabitans daechungensis [111] with 92.37% similarity in the 16S rRNA gene. Therefore, OSBR104 may represent a novel taxon (Fig. 4). Additionally, isolates related with Microbacterium amylolyticum (C.AS10 and C.AW5), Microbacteium ginsengiterrae (C.AW1), Micrococcus luteus (C.OS4) and Nocardioides salarius (OSBR100) showed low similarities with the mentioned strains (Supplementary Table 2) and may represent new taxa (Fig. 4).
Both classes of the phylum Planctomycetota, namely Planctomycetia and Phycispharae were isolated in this study. Planctomycetia isolates obtained were affiliated with genera Alienomonas (1 isolate), Rhodopirellula (2 isolates) and Maioricimonas (1 isolate) (Fig. 2). Four out of the 5 Planctomycetia isolates showed only up to 97.95% similarity in the 16S rRNA gene with the closest related described species (Fig. 4; Supplementary Table 2). These data are indicative of novel Planctomycetia taxa (Fig. 4). Within the less known Planctomycetota class Phycispharae, only one isolate (ASED31) was obtained, for which the species Algisphaera agarilytica was the most related one but in a far distant level (88.8% similarity in the 16S rRNA gene), being putatively indicative of a new family within Phycispharae (Fig. 4). Curiously, all planctomycetal isolates obtained in the present study were obtained from Aveiro saltern.
Within Rhodothermota, the family Balneolaceae was the only one recovered with 3 isolates (ASBR13, AS101 and OSBR103), that due to the phylogenetic distance, could not be related with any known Rhodothermota genus (Fig. 4).
Inside the phylum Gemmatimonadota, only one isolate (AW12) phylogenetically placed within the family Longimicrobiaceae was retrieved (Fig. 3). However, the 16S rRNA gene distant relatedness with the closest described species, represented by 85.82% similarity with Longimicrobium terrae, may be indicative of a novel family (Fig. 4).
The most frequent fungal genus was Penicillium, since 26.6% of the overall microbial isolates were classified within this genus (Supplementary Table 2). The most common bacterial genera retrieved from both salterns were the Actinomycetotal Streptomyces (8.4%) and the Pseudomonadota Rhodovibrio (7.1%), followed by the genus Sinomicrobium (6.5%) that is a member of phylum Bacteroidota (Supplementary Table 2).
Regarding overall phyla and genera, the isolated microorganisms associated with
Aveiro saltern showed a higher diversity (Fisher’s
| Fisher’s |
Margalef’s | Simpson’s | |||||
| Aveiro | Olhão | Aveiro | Olhão | Aveiro | Olhão | ||
| General by phylum | 2.389 | 1.74 | 1.733 | 1.259 | 0.1983 | 0.2624 | |
| (2.04–2.389) | (1.74–1.74) | (1.517–1.733) | (1.259–1.259) | (0.1756–0.2365) | (0.2218–0.3471) | ||
| General by genus | 17.06 | 12.85 | 6.934 | 5.037 | 0.07597 | 0.1826 | |
| (12.82–17.06) | (6.969–12.85) | (5.85–6.934) | (3.526–5.037) | (0.05715–0.1071) | (0.1136–0.3001) | ||
| Fungi | 1.841 | 0.2185 | 1.228 | 0 | 0.6095 | 1 | |
| (0.8763–1.841) | (0.2185–0.2185) | (0.6139–1.228) | (0.4172–0.855) | ||||
| Ascomycota | 1.344 | 0.2185 | 0.932 | 0 | 0.6576 | 1 | |
| (0.5116–1.344) | (0.3107–0.932) | (0.4752–0.8528) | |||||
| Basidiomycota | 0 | – | 0 | – | 1 | – | |
| Bacteria | 16.21 | 22.82 | 6.254 | 5.482 | 0.06453 | 0.07031 | |
| (11.32–16.21) | (9.49–22.82) | (5.096–6.254) | (3.751–5.482) | (0.05209–0.08656) | (0.06641–0.1309) | ||
| Bacillota | 4.632 | 5.403 | 1.82 | 2.085 | 0.2346 | 0.2727 | |
| (1.576–4.632) | (1.359–5.403) | (0.9102–1.82) | (0.8341–2.085) | (0.2099–0.5062) | (0.1901–0.5702) | ||
| Pseudomonadota | 5.949 | 3.878 | 2.762 | 1.542 | 0.1627 | 0.3061 | |
| (3.143–5.949) | (0.9354–3.878) | (1.842–2.762) | (0.5139–1.542) | (0.1302–0.2899) | (0.2653–0.7551) | ||
| Bacteroidota | 0.7972 | 0.7959 | 0.4551 | 0 | 0.8025 | 1 | |
| (0.5556–0.8025) | |||||||
| Actinomycetota | 1.968 | 13.19 | 1.276 | 2.919 | 0.2968 | 0.157 | |
| (1.399–1.986) | (2.261–13.19) | (0.9568–1.276) | (1.251–2.919) | (0.2401–0.4669) | (0.1405–0.3554) | ||
| Planctomycetota | 9.284 | – | 1.864 | – | 0.28 | – | |
| (1.235–9.284) | (0.6213–1.864) | (0.28–0.52) | |||||
| Rhodothermota | 0.7959 | 0 | 0 | 0 | 1 | 1 | |
| Gemmatimonadota | 0 | – | 0 | – | 1 | – | |
In Aveiro, the highest diversity and richness were registered within Planctomycetota and Pseudomonadota, while the lowest value of dominance was observed in Pseudomonadota, highlighting the heterogeneity of the Pseudomonadota isolates community (Table 2). At Olhão saltern, Actinomycetota and Pseudomonadota showed the highest diversity, while the lowest diversity was observed within Ascomycota and Rhodothermota (Table 2). Additionally, in Olhão, the highest values of diversity and richness along with the lowest value of dominance were observed within Actinomycetota (Table 2), demonstrating the high heterogeneity of Actinomycetotal isolates obtained.
Despite the high isolated diversity, the Mao’s Tau rarefaction curve presented a continuous rise without achieving an asymptote, confirming that not all microbial diversity was recovered (Fig. 5).
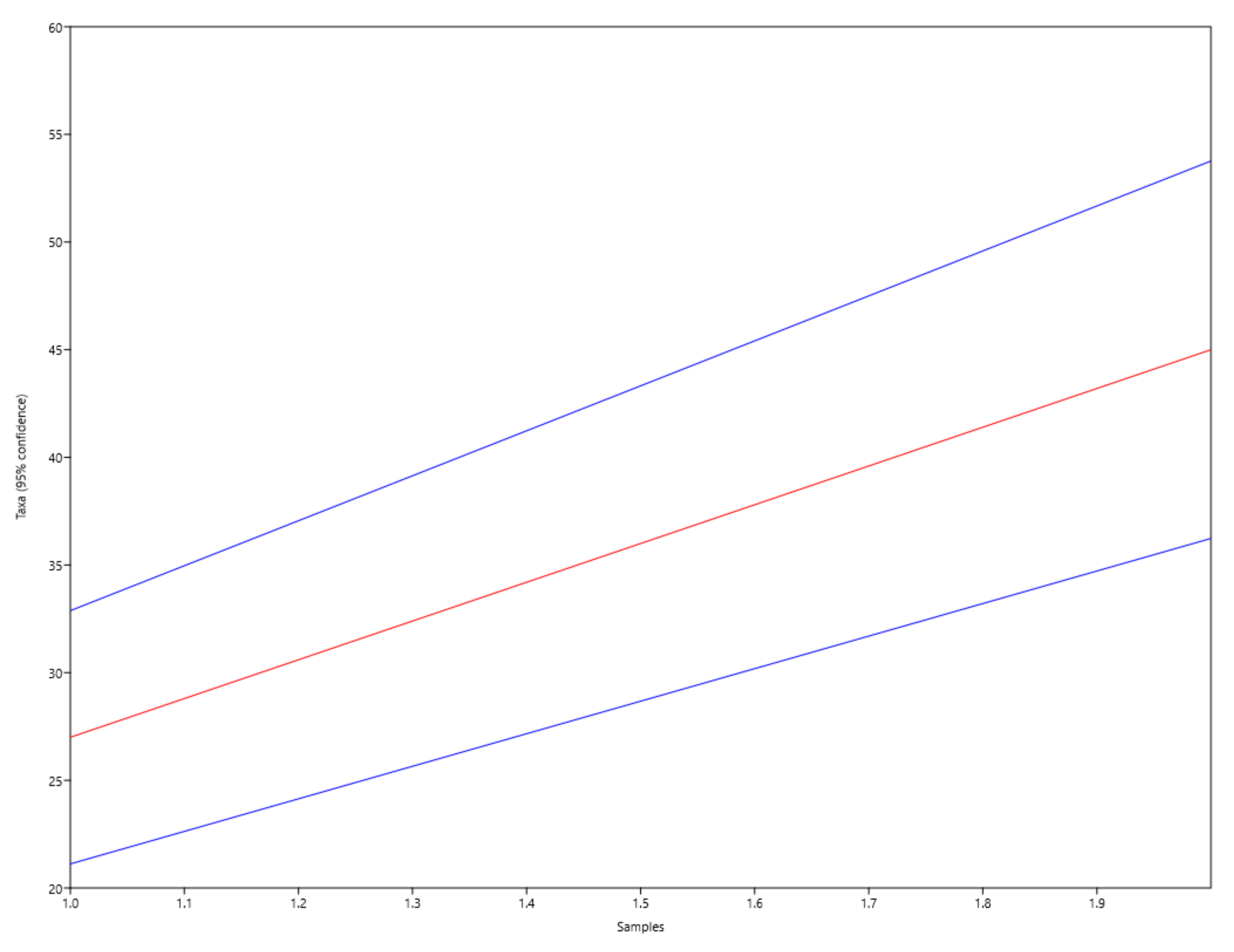 Fig. 5.
Fig. 5.Mao’s Tau rarefaction curve (red) obtained on the genera diversity analysis of microbial isolated communities from Aveiro and Olhão salterns and representing the species accumulation with 95% confidence intervals (in blue).
Concerning the fungal biodiversity, 3 singletons were observed in Aveiro, while none was detected in Olhão. These singletons represent 6.4% of the whole fungal diversity (Supplementary Material). The overall bacterial diversity was comprised by 40 different genera, with 25 singletons, that represent 23.4% of the overall bacterial diversity (Supplementary Table 2). These may be rare members of the isolated bacterial communities of salterns. Additionally, from the 28 bacterial genera recovered from Aveiro saltern, 12 occurred as singletons, representing 16.0% of the overall diversity of isolates, while Olhão taxa biodiversity consisted in 20 different bacterial genera, where 13 occurred as singletons, representing 37.1% of the overall diversity of isolates (Supplementary Table 2). From all these singletons, only 2 were common to both salterns, namely, the Bacillota genera, Mesobacillus and Rossellomorea (Supplementary Table 2).
The Sørensen and Bray-Curtis indexes were determined between the microbial populations of both salterns as 0.33 and 0.43, respectively. These parameters are measures of the similarity between both microbial populations, therefore highlighting the dissimilarities between them. Taking into account that sampling dates were close and that all the procedures undertaken were the same for all samples, these differences between the two microbial populations studied may be a result of the different geographical locations of the two salterns (Aveiro at the North of Portugal vs. Olhão at the South of Portugal).
The biotechnological potential of all isolates obtained in this study was screened by the PCR analysis of PKS-I and NRPS genes (Table 3; Supplementary Table 2). From the 154 microorganisms isolated in this study, 51 (33.1%) did not present any PKS or NRPS genes and 80 (52%) presented one (64–41.6% PKS positive and 16–10.4% NRPS positive) or both genes (23 bacteria, 14.9%) (Table 3; Supplementary Table 2). Overall, this screening revealed that 66.9% of the isolates may present at least one of the genes, revealing their putative biotechnological potential (Table 3; Supplementary Table 2). Although microbial ability to produce bioactive metabolites is not strictly linked to PKS and NRPS genes, because many other genes have been associated with bioactive NPs production [112], the genomic presence of these genes already proved to be a good indicator of the production of antimicrobial compounds [113]. Furthermore, NPs derived from these genes, as polyketides (PK), nonribosomal peptides (NRP) or even PK/NRP hybrids, have demonstrated their high economical and pharmacological value [20, 114]. These gene clusters were previously detected in ascomycetes and in a high number, which may be associated with the production of several different NRPS- and PKS-derived NPs [115].
| Phylum | Genus | Number of Isolates | Only NRPS | Only PKS | Both |
| Actinomycetota | Streptomyces | 13 | 0 | 9 | 1 |
| Nocardiopsis | 6 | 0 | 6 | 0 | |
| Micrococcus | 1 | 0 | 0 | 1 | |
| Microbacterium | 5 | 1 | 0 | 2 | |
| Brevibacterium | 2 | 0 | 1 | 0 | |
| Rothia | 2 | 0 | 1 | 0 | |
| Gordonia | 1 | 0 | 1 | 0 | |
| Brachybacterium | 1 | 0 | 0 | 0 | |
| Nocardioides | 1 | 0 | 0 | 0 | |
| Tsukamurella | 1 | 0 | 0 | 1 | |
| Iamiaceae* | 1 | 0 | 0 | 0 | |
| Pseudomonadota | Rhodovibrio | 11 | 1 | 3 | 1 |
| Wenzhouxiangella | 4 | 1 | 2 | 1 | |
| Microbulbifer | 4 | 0 | 1 | 3 | |
| Luteimonas | 3 | 1 | 0 | 1 | |
| Halomonas | 3 | 0 | 3 | 0 | |
| Marinobacter | 2 | 1 | 0 | 0 | |
| Alcanivorax | 2 | 0 | 0 | 0 | |
| Brevundimonas | 1 | 1 | 0 | 0 | |
| Stappia | 1 | 0 | 0 | 1 | |
| Roseibacterium | 1 | 0 | 0 | 0 | |
| Aureimonas | 1 | 0 | 1 | 0 | |
| Salinicola | 1 | 0 | 1 | 0 | |
| Bacillota | Bacillus | 3 | 3 | 0 | 0 |
| Metabacillus | 5 | 0 | 0 | 0 | |
| Alkalihalobacillus | 2 | 0 | 2 | 0 | |
| Paenibacillus | 2 | 0 | 0 | 2 | |
| Thalassobacillus | 2 | 0 | 0 | 1 | |
| Paenisporosarcina | 1 | 1 | 0 | 0 | |
| Mesobacillus | 2 | 1 | 0 | 0 | |
| Cytobacillus | 1 | 0 | 0 | 0 | |
| Rossellomorea | 2 | 2 | 0 | 0 | |
| Bacteroidota | Sinomicrobium | 10 | 0 | 8 | 0 |
| Psychroflexus | 1 | 0 | 0 | 1 | |
| Planctomycetota | Alienimonas | 1 | 0 | 0 | 0 |
| Maiorcimonas | 1 | 1 | 0 | 0 | |
| Rhodopirellula | 2 | 0 | 1 | 1 | |
| Algisphaera | 1 | 0 | 1 | 0 | |
| Rhodothermota | Balneolaceae* | 3 | 0 | 1 | 2 |
| Gemmatimonadota | Longimicrobiaceae* | 1 | 0 | 0 | 0 |
| Ascomycota | Penicillium | 41 | 1 | 21 | 4 |
| Cladosporium | 3 | 0 | 1 | 0 | |
| Aureobasidium | 1 | 1 | 0 | 0 | |
| Alternaria | 1 | 0 | 0 | 1 | |
| Basidiomycota | Sporobolomyces | 1 | 0 | 0 | 0 |
The high percentages and widespread distribution of NRPS and PKS genes observed in the phyla Pseudomonadota, Actinomycetota, and Bacillota was already reported by Wang et al. [115]. In contrast, both Bacteroidota and Planctomycetota are less explored phyla in what concerns their potential for the production of bioactive molecules, being research fields that are being launched with encouraging results obtained from a preliminary screening of NRPS and PKS genes [116, 117, 118]. Concerning Rhodothermota, since this phylum was recently branched out from Bacteroidota [8], no studies targeting their overall genomic potential for production of bioactive NPs are yet published. Regardless of the only 3 representatives obtained, since, up to the present moment, the phylum Rhodothermota has only 13 described species from 4 different families of only 1 class, the presence of at least one of these genes in all isolated members of this phylum pave the way for future deeper studies on this poorly known phylum. As detected in the present work, NRPS and PKS-I gene clusters were previously detected in ascomycetes and in a high number, which may be associated with the production of several different NRPS- and PKS-derived NPs [115]. No NRPS or PKS-I amplicons were detected either in Gemmatimonadota or Basidiomycota (Table 3; Supplementary Table 2). In fact, Gressler et al. [119] reported that PKSs and NRPSs are not the main contributors for basidiomycetes diversity of NPs. On the other hand, the poorly known phylum Gemmatimonadota was firstly described less than two decades ago and presently only has 6 described species. This scarce number of described species is due to the difficulties in successfully isolating Gemmatimonadota members in laboratory, despite being widely distributed in the environment [120]. Therefore, most of the research made on its bioactive potential relies on metagenomes, which demonstrated the presence of not only NRPS, PKS and hybrid PKS/NRPS gene clusters [121], but also a widespread and high prevalence of bacteriocin-related biosynthetic gene clusters [122].
Overall, the microbiota isolated from both salterns showed a great biotechnological potential. Curiously, although with lower number of isolates, the phyla that showed higher bioactive potential were the Rhodothermota, Bacteroidota and Planctomycetota, and not the well-known bioactive top producers, Actinomycetota [123].
This work provides evidence that salterns remain an understudied extreme environment and that culturomic methods are still an important approach for the study of microbial diversity, since a high number of novel taxa from different phyla was obtained. In fact, although metagenomics is a fundamental approach for novel microbial diversity detection, pure cultures are needed to enable the biological characterization of many species yet unknown. Furthermore, the isolated microbiota from salterns showed a substantial underexplored bioactive potential providing data and biological material that encourages further research works as bioactivity screenings.
In general, the overall microbial diversity obtained has been previously associated with salterns or with other hypersaline environments, however our results also pointed out genera not yet linked to these environments.
NP, natural product; PKS-I, polyketide synthase I; NRPS, nonribosomal peptide synthethase; SW, seawater; PCR, polymerase chain reaction; NCBI, National Centre for Biotechnology Information; RT, room temperature; SCN, starch-casein-nitrate agar; NPS, nutrient-poor sediment extract agar.
EA, MFC and OML designed the research study. EA performed the research. EA, MFC and OML analyzed the data. EA, MFC and OML wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors EA, MFC and OML thank to national funds provided by the Fundação para a Ciência e Tecnologia (FCT; Foundation for Science and Technology) at Portugal, and European Regional Development Fund (ERDF) and COMPETE under the project INNOVMAR—Innovation and Sustainability in the Management and Exploitation of Marine Resources, reference NORTE-01-0145-FEDER-000035, Research Line NOVELMAR. EA was supported by FCT for the Ph.D. Grant SFRH/BD/125527/2016. MFC wishes to acknowledge CEEC program supported by FCT (CEECIND/02968/2017), Fundo Social Europeu and Programa Operacional Potencial Humano. This work was supported by the Strategic Funding UIDB/04423/2020 and UIDP/04423/2020 through national funds provided by FCT and ERDF. This research was partially supported by the Strategic Funding UID/Multi/04423/2019 through national funds provided by FCT and ERDF, in the framework of the program PT2020, and by the Project Funding NORTE-01-0145-FEDER-000035, designed ATLANTIDA – Plataforma para a Monitorização do Oceano Atlântico Norte e Ferramentas para a Exploração Sustentável dos Recursos Marinhos, provided by FCT and ERDF.
The authors declare no conflict of interest.
