†These authors contributed equally.
Academic Editor: Michael H. Dahan
Background: Endometrial cancer (EC) is one of the most common gynecologic cancers of the female reproductive system. Its incidence and mortality are currently increasing. Patients with early-stage EC have a much better prognosis than those with late-stage EC. Therefore, early detection, diagnosis, and treatment are critical to improving the outcome for EC patients. The proposition of molecular classification promotes the individualization for diagnosis and treatment of EC. TCOF1 has been identified as an oncogenic gene in several tumors but has been seldom studied in EC. Methods: TCGA and immunohistochemistry (IHC) experiments were performed to verify the protein level of TCOF1 expressed in endometrial cancer while its prognostic ability in EC patients was assessed by the TCGA database. Linked Omics database, Web Gestalt gene enrichment, and string database were applied to analyze the possible biological functions of TCOF1 in EC. Mutation types of TCOF1 in EC and its mutation frequency were explored in c-BIOPORTAL. The Relationship between molecules was detected by utilizing the GEPIA database. Results: TCOF1 is up-regulated in endometrial cancer compared to para cancer and it was positively correlated with poor prognosis of patients. TCOF1 is mutated in endometrial cancer and is closely associated with microsatellite instability (MSI), this being one type of molecular classification in EC. Conclusions: TCOF1 may function as a potential biomarker and is associated with molecular classification in endometrial cancer.
Endometrial cancer (EC) is the leading malignancy of the female reproductive system in developed countries. There were 417,000 new cases of endometrial cancer and 97,000 deaths reported in 2020 [1, 2]. Hysterectomy and or bilateral salpingo-oophorectomy are the standard treatments for endometrial cancer. However, it is well documented that patients diagnosed at early-stage EC experienced a better prognosis after surgical treatment or combined radiotherapy than those diagnosed at a late stage [3]. It is generally accepted that causative factors of endometrial cancer include obesity, long-term stimulation of estrogen, diabetes mellitus, and delayed menopause [4, 5]. However, the pathogenesis of endometrial cancer is still unclear. New biological markers for endometrial cancer are being sought. Molecular classification has been proposed by The Cancer Genome Atlas (TCGA) for the diagnosis of endometrial cancer, which classified endometrial cancer into 4 types, including POLE mutation, low copy number, high copy number, and microsatellite instability (MSI). The incorporation of TCGA into diagnostic guidelines suggests that it is of importance for individualized diagnosis and treatment of patients with endometrial cancer [6, 7, 8, 9].
Lynch syndrome is a familial genetic disorder and is a well-known risk factor for endometrial cancer [10]. The probability of developing endometrial cancer in patients with Lynch syndrome is comparable to that of colorectal cancer [11]. As cancer often occurs in Lynch syndrome, this subset makes up 2% of patients with endometrial cancer. Approximately 50% of patients with Lynch syndrome will develop endometrial cancer [12].
Mutations in DNA mismatch repair genes are an important factor in the pathogenesis of Lynch syndrome [10, 13]. Utilized in the detection for Lynch syndrome, assessment of DNA mismatch repair (MMR) proteins by IHC is a part of the diagnostic evaluation for patients with EC [14]. The primary function of MMR is to correct mispairing individual nucleotides during DNA replication and mutations in MMR cause microsatellite instability. MLH1, MSH2, MSH6, and PMS2 are the four most commonly mutated mismatch repair genes [15, 16].
TCOF1 (Treacle Ribosome Biogenesis Factor 1) is localized on chromosome 5 (5q32-33.3) with 152 KD. It was initially identified as a gene closely associated with the pathogenesis of Treacher Collins syndrome [17]. Further findings demonstrated that TCOF1 is involved in the development of multiple diseases and has important roles in cell proliferation, apoptosis, and DNA damage repair [18]. Upregulation of TCOF1 promotes the growth and stemness formation in breast cancer [19]. TCOF1 activates the KRAS gene and EMT signaling pathway, while a negative relevance exists between TCOF1 and anti-tumor immune cell infiltration in hepatocellular carcinoma [20]. As a potential oncogene in hepatocellular carcinoma, patients with higher expression of TCOF1 may experience a worse prognosis. TCOF1 encodes a cytoplasmic protein that is involved in the transcription of ribosomal DNA which is specifically enriched in telomeres, leading to defective telomere replication and genomic instability [21]. This has given rise to the study of TCOF1 in endometrial cancer patients.
In this research, we evaluated the expression of TCOF1 in patients with EC and explored its relationship with clinicopathologic characteristics by public databases and IHC. To detail the value of TCOF1 in EC, we investigated the relationship between TCOF1 and MMR (MLH1, MSH2, MSH6, and PMS2), which is related to microsatellite instability (MSI) [22], one kind of molecular classification of EC as TCOF1 is involved in DNA damage repair [23]. To the best of our knowledge, MMR is a response to DNA damage [24].
The UALCAN (http://ualcan.path.uab.edu/) database, which can be can be used to analyze the differential expression of genes in normal tissues and tumor tissues and explore the relationship between genes and clinicopathologic parameters of patients, is an online database based on the TCGA database. We explored the expression of TCOF1 in pan-cancer and analyzed the correlation between TCOF1, NOLC1, MLH1, MSH2, MSH6, and PMS2 and their clinicopathologic parameters in patients with endometrial cancer. NOLC1 plays an important role in nucleolar and rRNA synthesis and participates in ribosomal biogenesis [25]. Studies have demonstrated that it can promote tumor proliferation, invasion, and metastasis [25, 26]. Also indicated is that NOLC1 and TCOF1 are mineralocorticoid receptor-related proteins and can be used as regulatory cofactors of CK1 [27, 28]. MLH1, MSH2, MSH6, and PMS2 play an essential role in mismatch repair (MMR) [29].
TCGA (The Cancer Genome Atlas program) was jointly launched by NCI (National Cancer Institute) and NHGRI (Human Genome Research Institute) in 2006 [30]. TCGA has tested 33 tumors from 20,000 patients, including 10 rare tumors, with a data volume of 2500t. Based on large-scale sequencing technology and through extensive cooperation, we can understand the molecular mechanism of cancer and finally get a complete set of “maps” related to all cancer genome changes, and improve scientific understanding of the molecular mechanism of cancer pathogenesis and improve the ability for diagnosis, treatment, and prevention. The database includes biospecimen, clinical information, sequencing reads, transcriptome profiling, simple nucleoside variation, copy number variation, and DNA methylation of patients. The TCOF1 mRNA expression data in the endometrial cancer dataset were downloaded from the TCGA database, which includes 543 endometrial cancer tissue specimens and 35 normal endometrial tissue specimens (23 pairs of cancer and para cancer tissue samples).
GEPIA is a database providing functionalities according to datasets from TCGA and GTEx [31], which help analyze gene expression correlations. We analyzed the correlation between TCOF1 and associated genes, including ARRB1, NOLC1, SNK2A2, KBTBD8, ARRB2, MSH2, MSH6, PMS2, and MLH1 in GEPIA.
c-Bioportal (http://www.cbioportal.org/) database provides the frequency and type of gene mutations. We analyzed that of TCOF1 in endometrial cancer patients.
String (https://cn.string-db.org/) is a database for analyzing protein interaction. Possible reciprocal proteins of TCOF1 were analyzed and the top 5 are shown.
Linkedomics database (http://www.linkedomics.Org/login.php) is a multidimensional online data analysis platform [32], which mainly consists of three analytical modules. The LinkFinder module is used to analyze the gene profiles co-expressed with TCOF1 in EC, and the LinkInterpreter module is used for GO annotation and KEGG enrichment analysis.
The endometrial cancer tissue microarray was obtained from Shanghai Outdo
Biotech Company. IHC test kit (PV-9000) was from Zsbio (Beijing, China)
for TCOF1 protein expression analysis. TCOF1 was incubated at a
concentration of 1:100 (PTG). IHC assay was performed as per our previous
research [33]. The final IHC score was determined by two senior
clinicopathologists, with a score of
t-test or Wilcoxon rank-sum test was performed to analyze the
differences between the two groups. Survival analysis was performed by
Kaplan-Meier. The diagnostic efficacy of the index was evaluated by the operating
characteristic curve (ROC). Spearman correlation analysis was used to assess the
correlation between TCOF1 and other genes in GEPIA database. Univariate
and Cox regressions were used to examine relationships between single
clinicopathologic parameters and clinical prognosis in patients with EC.
p
First, we explored the expression of TCOF1 in pan-cancer in the UALCAN
database which revealed that TCOF1 is up-regulated in most tumors
compared with normal tissues (Fig. 1a). Thus, we downloaded the data for
endometrial cancer from the TCGA database, which contained 543 cancer tissues and
35 normal tissues, including 23 pairs of carcinoma and its para cancer tissues.
This demonstrated that expression of TCOF1 mRNA was significantly higher
in EC tissues than that in normal groups (p
 Fig. 1.
Fig. 1.Expression of TCOF1. (a) Expression of TCOF1
in Pan-cancer in UALCAN. (b) Expression of TCOF1 in endometrial
carcinomain TCGA database based on unpaired tissue. (c) Expression of
TCOF1 in endometrial carcinomain TCGA database according to paired
tissue. (d) The typical image of TCOF1 in EC (scale bar: 50
Further subgroup analysis of multiple clinicopathologic characteristics in 546
cases of endometrial cancer and 35 cases of non-cancer via the online UALCAN
database was performed. This suggested that TCOF1 was highly expressed
in subgroups, including tumor stage, patient race, patient weight, patient age,
menstrual status, historical subtypes, and P53 mutation status than that
of normal tissue (Fig. 2). Considering TCOF1 may act as a potential
biomarker for the diagnosis of endometrial cancer, we explored the relationship
between TCOF1 and the prognosis of patients with EC, which demonstrated
that patients had worse overall survival, disease-free survival, and
progression-free survival with higher expression of TCOF1 (Fig. 3a–c).
As shown, AUC at 1-year, 3-year as well as 5-year is larger than 0.5. (Fig. 3d).
Univariate Cox regression indicated that TCOF1 was associated with Age
(p
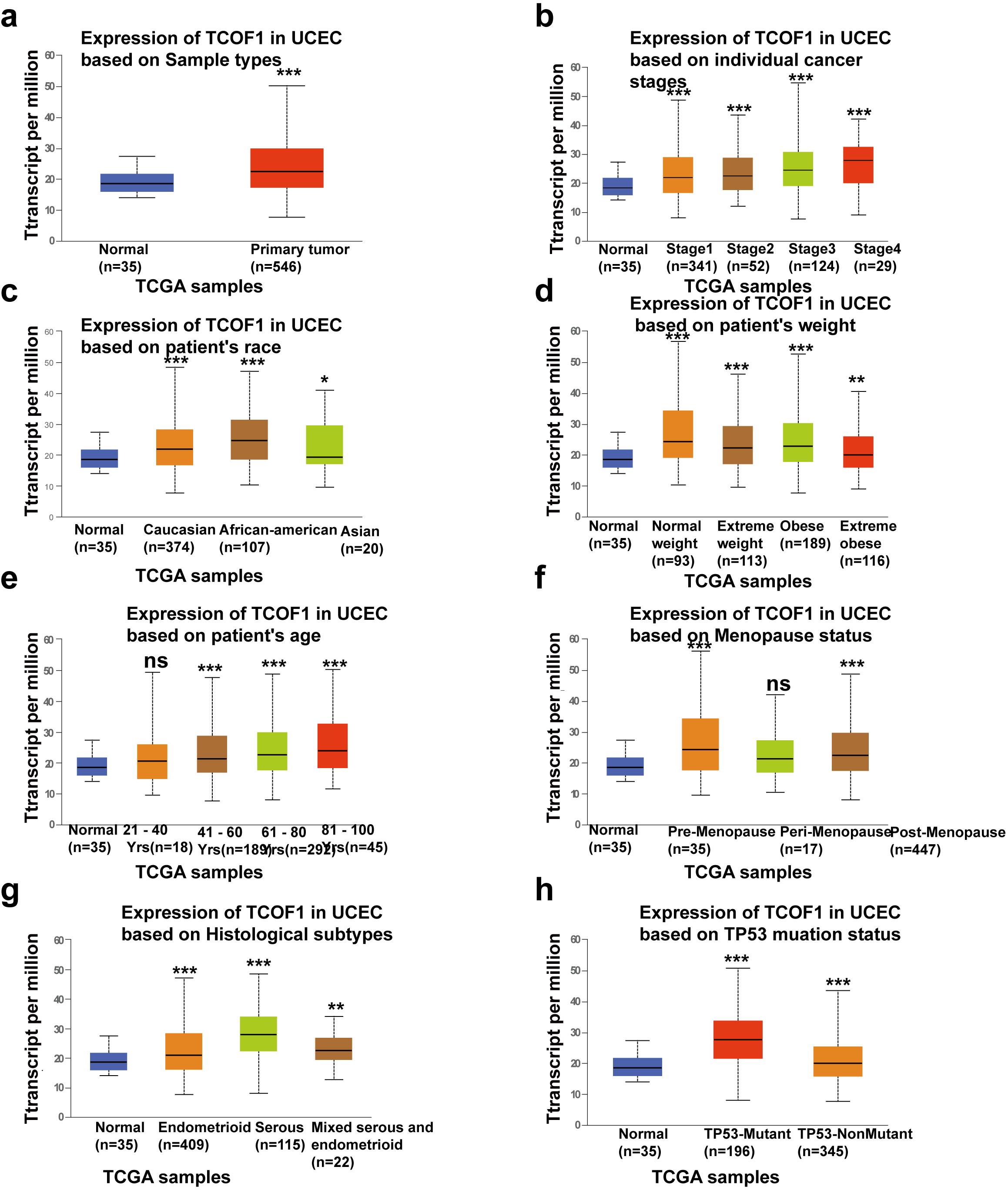 Fig. 2.
Fig. 2.Relationship between TCOF1 mRNA expression and clinical
features of EC. Analysis is shown in UALCAN database as sample types (a), cancer
stages (b), race (c), weight (d) and patient’s age (e), menopause status (f),
historical subtypes (g), and TP53 mutation status (h) respectively. *p
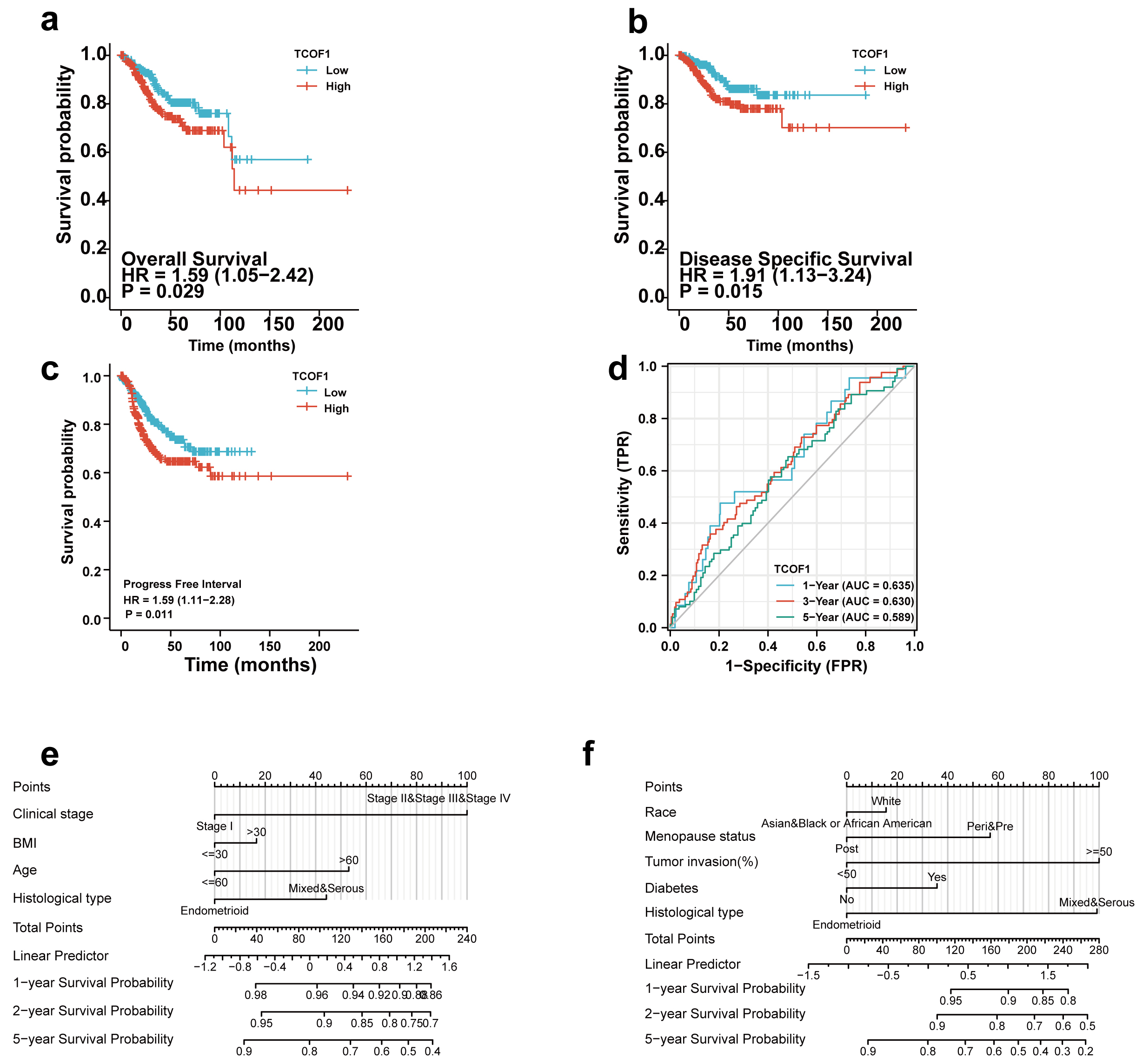 Fig. 3.
Fig. 3.Survival analysis for patients with endometrial carcinoma depending on TCOF1 mRNA level. TCGA cohorts are shown for OS (a), DSS (b), and PFI (c). ROC curve of TCOF1 gene expression in patients with EC (d). Univariate Cox regression results based on TCGA database (e, f).
Protein-Protein Interaction Networks (PPI) are defined as protein-protein interaction to participate in biological processes such as cell energy metabolism, cell cycle regulation, and intracellular signal pathway [34]. The co-expressed genes of TCOF1 were enriched and analyzed by LinkOmics. It showed that numerous genes were closely associated with TCOF1 (Fig. 4a). Graphs showed the top 50 significant genes which are associated with TCOF1 (Fig. 4b,c). String database is a powerful database for protein interactions. The top 5 interactants of TCOF1, which includes CSNK2A2, NOLC1, KBDBC8, ARRB1, ARRB2 are shown in Fig. 5a. We examined thecorrelation between TCOF1 and CSNK2A2, NOLC1, KBDBC8, ARRB1, ARRB2 expression in endometrial cancer in the GEPIA database. TCOF1 was positively correlated with CSNK2A2, NOLC1, KBDBC8, ARRB2 expression (Fig. 5c–f ), but not ARRB1 (Fig. 5b). It was obvious that TCOF1 was more closely aligned with NOLC1, and the study suggested that TCOF1 and NOLC1 were paralogous homologs [27]. Therefore, we investigated the clinicopathologic parameters according to the expression of NOCL1 in endometrial cancer via UALCAN database. Interestingly, NOLC1 was closely associated with tumor stage, patient race, patient weight, patient age, menstrual status, tissue classification, and P53 mutation status in endometrial cancer patients, which is similar to that of TCOF1 (Fig. 6).
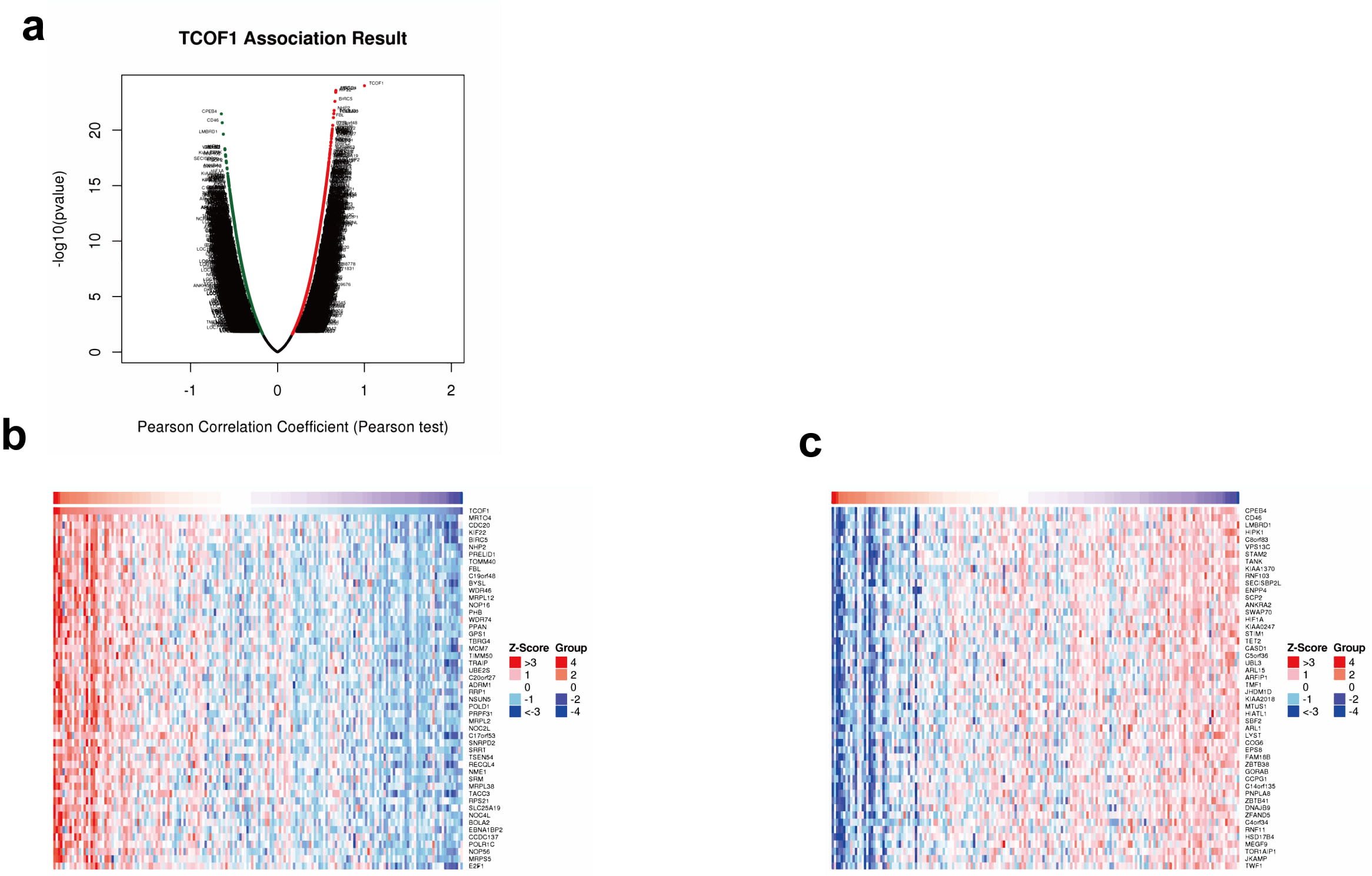 Fig. 4.
Fig. 4.Co-expression genes of TCOF1. TCOF1 gene co-expression gene volcano map (a) in LinkOmics. Heat map of top 50 most significantly positively (b) and negatively (c) associated genes co-expressed with TCOF1.
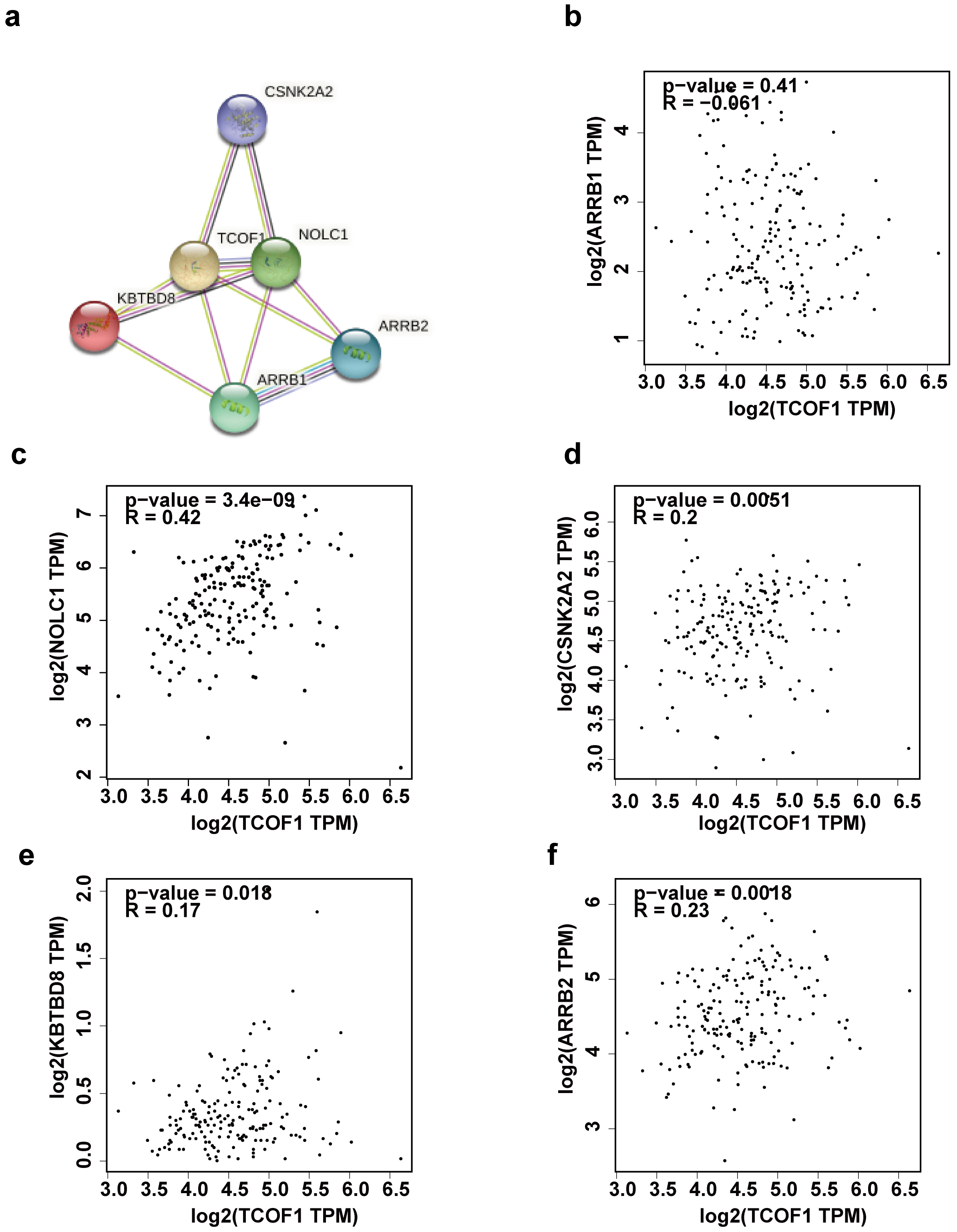 Fig. 5.
Fig. 5.Protein-Protein Interaction Networks of TCOF1. The top 5 interactants of TCOF1 in String database (a). Scatter diagram evaluating co-relationship between TCOF1 and ARRB1 (b), NOLC1 (c), SNK2A2 (d), KBTBD8 (e), ARRB2 (f) in GEPIA database.
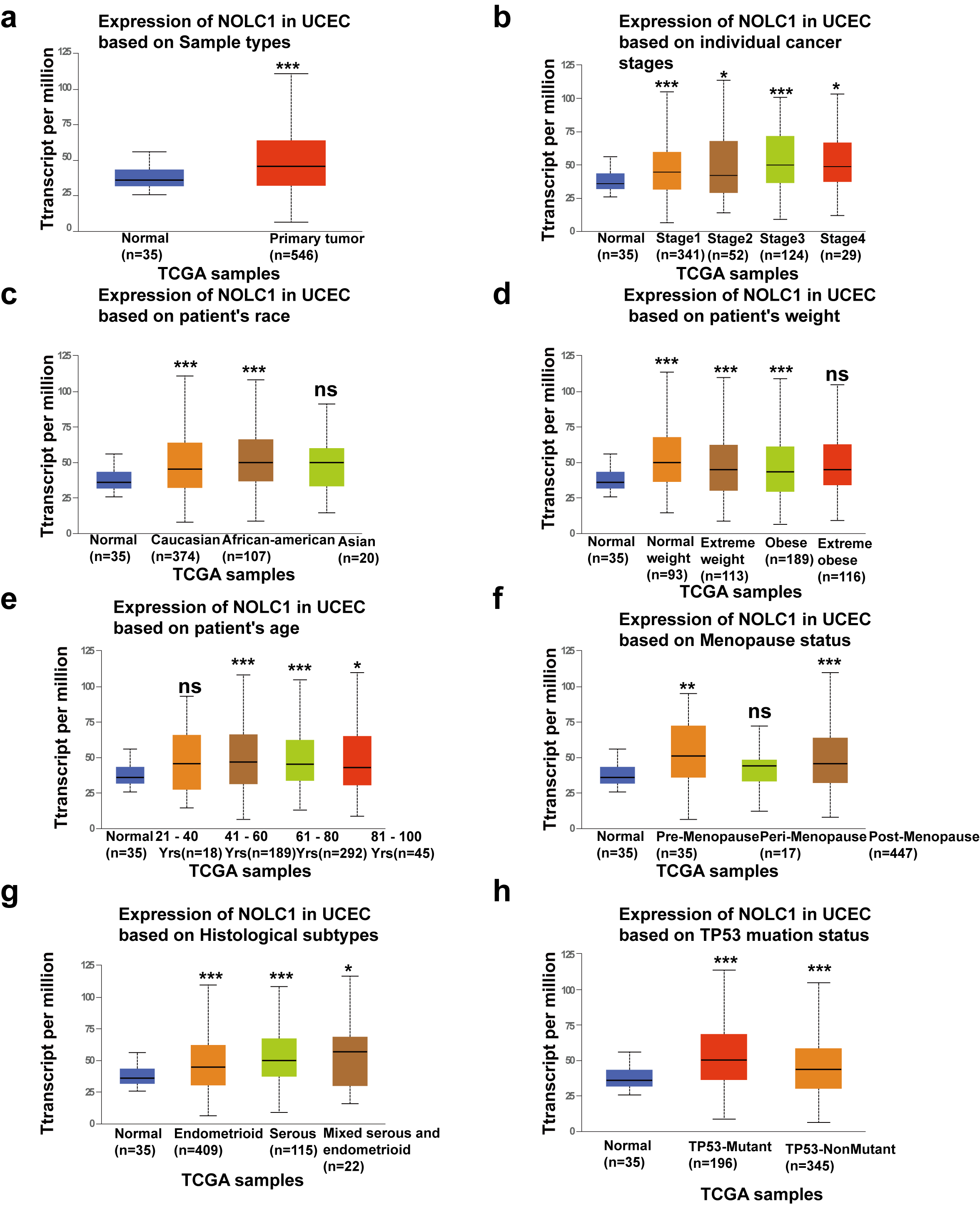 Fig. 6.
Fig. 6.Box diagram shows the expression of NOLC1 among
different subgroups. Sample types (a), individual cancer stages (b), race (c),
weight (d), age (e), menopause status (f) historical subtypes (g) and TP53
mutation (h). respectively. *p
To clarify the possible molecular mechanism involved in the process that TCOF1 promotes endometrial cancer progression, enrichment analysis was applied to determine the co-expressed genes of TCOF1. Gene ontology (GO) enrichment analysis was divided into three sections, which contained biological processes, cellular components as well as molecular functions. Genes co-expressed with TCOF1 were chiefly involved in biological regulation, the process of metabolism, and reaction to stimulus. Cellular component analysis revealed that the genes co-expressed with TCOF1 were mainly located in the membrane, nucleus, and membrane-enclosed lumen (Fig. 7).
 Fig. 7.
Fig. 7.Functional enrichment analysis of TCOF1 co-expressed genes by gene ontology (GO) enrichment. (a) Biological process. (b) Cellular component. (c) Molecular function.
Given that mutations in TCOF1 are an important cause of Treacher
Collins Syndrome [35], to obtain knowledge of the mutation of TCOF1 in
EC, c-BioPortal database was searched to investigate the mutation of
TCOF1. Results showed that among 242 patients with endometrial cancer, 8
patients had TCOF1 mutations, including 7 missense mutations and 1
truncating mutation(Fig. 8a,b). Being that TCOF1 participates in the
process of DNA damage repair with DNA mismatch repair is a measure to deal with
DNA damage repair [36], and that deletion of DNA MMR causes the gathering of
mismatches within the cycle of DNA replication, resulting in the development of
microsatellite instability (MSI) [37]. MLH1, MSH2,
MSH6, and PMS2 are the typical genes of DNA mismatch repair. We
explored the relationship between TCOF1 and the genes of DNA mismatch
repair in the GEPIA database. The results showed that MLH1 (R = 0.12,
p
 Fig. 8.
Fig. 8.Mutation of TCOF1 and its relationship with typical molecular of MMR. TCOF1 specific mutation site in the c-BioPortal database (a). Scatter diagram evaluating correlation ship between TCOF1 and MSH2 (b), MSH6 (c), PMS2 (d), MLH1 (e) in GEPIA database.
Endometrial cancer was classified into 4 types according to histological types, including endometrioid adenocarcinoma, plasmacytoma, clear cell-like adenocarcinoma of the uterus, and mixed type [38]. The risk factors that predispose to endometrial cancer and the clinical outcomes of endometrial cancer patients are also diverse [4], which suggests that endometrial cancer is a heterogeneous disease that requires more individualized diagnosis and treatment. The study clearly demonstrated that patients with early-stage endometrial cancer had a better prognosis than those with advanced recurrent metastatic disease after aggressive surgical treatment [3]. Therefore, it is extremely important to develop new therapeutic regimens and screen new biological markers to facilitate early diagnosis and personalized treatment for patients. A large set of new biomarkers was found with the advent of bioinformatics, which opened an era of big data screening for biomarkers in the field of oncology. TCOF1 gene mutation is closely associated with the pathogenesis of Treacher Collins syndrome [39], which is involved in cell proliferation, apoptosis, and DNA damage repair processes [18].
In this study, using the TCGA database, we found that TCGA mRNA showed high expression in endometrial cancer and that high expression of TCOF1 indicated worse OS, DFS, and DFP. To further validate this result, we performed an IHC assay to confirm the protein level of TCOF1 in endometrial cancer tissue. Results were consistent with the TCGA database. Subgroup analysis showed that the expression of TCOF1 in various clinicopathologic parameters was higher than that in the normal group. This suggests that TCOF1 may act as a potential diagnostic indicator for endometrial cancer. To explore the mechanism of TCOF1 involvement in endometrial carcinogenesis, gene co-expression was used to analyze the possible enrichment of TCOF1. PPI analysis revealed that TCOF1 may interact with NOLC1 which was reported to be a paralog of TCOF1, but it has not been studied in endometrial cancer. Thus we evaluated the relationship between NOLC1 and the clinical prognosis of patients with endometrial carcinoma with results being similar to TCOF1. It was revealed that the mutation of TCOF1 plays an important role in the diagnosis of Treacher Collins syndrome [17]. Thus, we evaluated this mutation in endometrial cancer. Considering that TCOF1 improves resistance to DNA damage response [23] and MMR belongs to the styles of DNA damage repair, we discussed the co-expression relationship with TCOF1 and MLH1, MSH2, MSH6, and PMS2, which are the typical genes in MMR. It was revealed that TCOF1 is positively associated with all of the MMR listed above. More importantly, the expression of MSH2, MSH6, and PMS2 was closely related to clinicopathologic parameters in endometrial cancer patients. Interestingly, microsatellite instability (MSI), which is one type of molecular classification of EC, is a signature feature of MMR [40].
We speculate that TCOF1 is of great significance in the progression of endometrial cancer through DNA damage repair mechanism and it may serve as a potential diagnostic criterion for microsatellite instability (MSI) endometrial cancer.
In conclusion, this study revealed the expression of TCOF1 in endometrial carcinoma. It is significantly up-regulated compared with normal tissues, which can be considered as a potential biomarker for the diagnosis and prognosis of endometrial cancer. In addition, through the analysis of the mutation and molecular expression correlation of TCOF1, it was found that TCOF1 is closely related to the typical molecules of MMR, one of the molecular classifications in EC. However, more in-depth cell experiments and clinical trials are needed to verify the value of TCOF1 in endometrial cancer.
TCOF1 may function as a potential biomarker in endometrial cancer and may be related to microsatellite instability (MSI), one of the molecular classifications in endometrial cancer.
SQG and HPJ design the study and were responsible for the conception of the present study. CQX and QHW analyzed the data and wrote the manuscript. CQX and QHW contribute the same efforts to this research. YLZ and QRG performed IHC assay and statistical analysis. All authors have read and approved the final manuscript.
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Shanghai Outdo Biotech Company (approval number: No.YB M-05-02).
Thanks to the team that established the public database, it provided free and convenient conditions for experimental exploration.
The present study was funded by The Nature Science Fund of Guangdong Province (grant no. 2020A15150).
The authors declare no conflict of interest.
