† These authors contributed equally.
Object: Long non-coding RNAs (lncRNAs) exert critical roles in cancer progression. However, the function of SNHG22 in epithelial ovarian carcinoma (EOC) still needs to be clarified. Methods: qRT-PCR was carried out to explore SNHG22 expression. Receiver operating characteristic (ROC) curve was used to evaluate the predictive ability. Glucose uptake and lactate secretion were used to monitor cancer cell glycolysis. Luciferase reporter and ChIP assays were conducted to investigate the molecular mechanism of SNHG22. Results: SNHG22 was found notably overexpressed in tumor tissues, cells and serum samples, which was regulated by SP1 with the stimulation of SNHG22 promoter activity. And SNHG22 expression in serum was an effective biomarker by ROC analysis. Moreover, SNHG22 facilitated glycolysis in EOC cells. Conclusions: Aberrant SNHG22 expression in serum could be used as a promising biomarker for EOC and SNHG22 promotes in glycolysis.
Epithelial ovarian carcinoma (EOC) is one of the most common malignant tumors. Globally, 1.2 million women suffered and 161 thousand died because of ovarian cancer in the year of 2015 [1]. EOC [2, 3, 4] is the seventh cancer and the eighth reason of death in female. And patients with EOC have a remarkable poor prognosis. This is frustrating that a large number of cases are diagnosed with advanced stages, mainly due to lacking significant early detecting method [5, 6]. Therefore, it is urgent for us to explore novel biomarkers for early detection, even potent therapies for EOC cases.
Long non-coding RNAs (lncRNAs) are described with more than 200 nucleotides and could not be translated into protein. Recently, lncRNAs [7, 8, 9, 10, 11] have been recognized as exerting numerous biological function in tumor progression. Aberrant expression of lncRNAs have been discovered in many kinds of malignant tumors. For instance, PCGEM1 is overexpressed and facilitates prostate cancer progression, for modulating cell proliferation [12]. MALAT1 is upregulated in hepatocellular carcinoma [13]. Although many ectopic lncRNAs expression have been found, their biological and genetic function is not fully illuminated in EOC progression. Hence, a suitable biomarker of lncRNAs needs to be cleared in EOC.
LncRNA SNHG22 is located in the 18q21.1 region of human genome. The precise biological role of SNHG22 have to illuminated in EOC. Our results indicated that SNHG22 expression level is overexpressed in cells, tissues and serum of EOC. Moreover, ectopic expression of SNHG22 predicts poor clinical outcomes. In addition, SNHG22 could modulate glycolysis. Additionally, SP1 adjusts SNHG22 transcription by means of binding to its promote. Hence, our findings propose that SNHG22 could be a promising therapy target in EOC.
Patients of EOC were enrolled from the liaoning cancer hospital between March
2010 and November 2015. The including criteria were as follow: patients were with
definite pathological diagnosis; And the exclusion criteria: patients were
received with chemotherapy or radiotherapy before surgery. Serum sample were
collected from EOC patients and healthy control. The tumor and paired non-tumor
tissues were also collected after lesion excision with 30 mins and stored in
liquid nitrogen, then transferred to -80
The EOC cell lines of ES-2, HO-8910, OVCAR-3, A2780 and human normal ovarian
epithelial (HNOE) were from American Type Culture Collection (ATCC). The test of
mycoplasma contamination was performed to make sure the cell lines were
mycoplasma-free. All the cells were cultured at 37
The expression levels of RNA were calculated by the qRT-PCR system. Total RNA
was extracted by TRIzol reagent (Invitrogen), and 1
The levels of glucose and lactate were calculated with a Glucose Colorimetric Assay Kit (BioVision, CA) and a Lactate Assay Kit (BioVision, CA) in line with the instructions of manufacturer.
ChIP was performed with the protocol of SimpleChIP®Enzymatic Chromatin IP Kit (Magnetic Beads) (#9003). Cells are fixed with formaldehyde and lysed, and chromatin is fragmented by partial digestion with Micrococcal Nuclease to obtain chromatin fragments of 1 to 5 nucleosomes. Chromatin immunoprecipitations are performed using ChIP-validated antibodies and ChIP-Grade Protein G Magnetic Beads. Anti-SP1 were performed in ChIP assay, and goat anti-rabbit IgG used as a negative control. After reversal of protein-DNA cross-links, the DNA was purified using DNA purification spin columns and was quantified by real-time qPCR analysis.
The SNHG22 promoter after PCR amplification was inserted into the downstream of the firefly luciferase gene in pGL3-Basic vector (Promega, USA) to construct pGL3-SNHG22 promoter reporter vector. Cells were co-transfected with a reporter construct (pGL3-basic plasmid or pGL3-SNHG22 plasmid) and shRNA or negative control. Dual Luciferase Reporter Assay System (Promega) was used to measure the luciferase activity 48 h after transfection. The relative luciferase activity was contrasted the Renilla luciferase and firefly luciferase activity.
All the data were showed as the mean
To investigate the expression status of SNHG22, we examined its expression using
qRT-PCR in a cohort of 60 paired and non-tumor tissues of EOC. The level of
SNHG22 was predominantly elevated in EOC tissues compared with the paired non-tumor tissues (Fig. 1A,B, P
 Fig. 1.
Fig. 1.SNHG22 is upregulated in EOC tissues, and cell lines.
(A, B) qRT-PCR analysis was used to evaluate SNHG22 expression in 60 paired
tumor and paired adjacent non-tumor tissues.
The expression profiles of SNHG22 in ES-2, HO-8910, OVCAR-3, A2780 and human
normal ovarian epithelial (HNOE) was detected with qRT-PCR.
*P
Our study also found that SNHG22 level in serum was remarkably upregulated in serum of EOC cases than healthy controls (Fig. 2A). Moreover, the AUC of ROC curve was 0.824 (95% CI 0.747–0.900), with the sensitivity 83.8% and specificity 70.9%, respectively (Fig. 2B). As a whole, our data show that SNHG22 is ectopic expressed in the circulation of EOC patients.
 Fig. 2.
Fig. 2.SNHG22 is upregulated in EOC serum.
qRT-PCR analysis was used to evaluate serum SNHG22 expression levels in EOC
patients and healthy controls.
ROC curve analysis of the diagnostic performance of serum SNHG22.
Kaplan-Meier survival curves revealed a relationship between SNHG22 expression
and overall survival of patients with EOC.
Comparison of the expression level of SNHG22 in serum among healthy controls,
early stage RCC patients, and late stage RCC patients.
Expression levels of SNHG22 in serum decreased after removal of primary tumors
in patients with EOC were quantified by quantitative RT-PCR (n = 24).
*P
Signaling pathways of downregulated genes after SNHG22 knockdown in A2780 cells
detected by microarray analysis.
Kaplan-Meier survival curves illustrated that high SNHG22 expression patients
had shorter overall survival time than low SNHG22 expression (Fig. 2C,
P
To investigate the biological function of SNHG22, we compared A2780 cells transfected with either sh-SNHG22 or a negative control. The pathway analysis showed that SNHG22 depletion had obvious effects on genes, which were significantly related to metabolic pathways (Fig. 2F). Accumulating evidence showed that cancer cells conduct glycolysis at a rapid speed than normal tissues [14]. And malignant tumor might be originated from abnominal energy metabolism. Consistent with previous reports, our data elucidated that glucose starvation could induce SNHG22 elevation and modulate the glycolytic metabolism in EOC cell. Glucose starvation obviously increased SNHG22 level in ES-2 cells (Fig. 3A). To explore the function of SNHG22 in glycolysis, we inhibited SNHG22 expression in A2780 cells and upregulated SNHG22 expression in ES-2 cells (Fig. 3B). Consequently, we calculated the extracellular acidification rate (ECAR) [15] to investigate whether SNHG22 could effectively influence glycolytic metabolism in EOC. Subsequently, sh-SNHG22 dramatically impaired ECAR levels in A2780 cells (Fig. 3C), compared to control group. We also explored the level of extracellular lactic acid, which is common used as a glycolysis metabolite. As a result, the production of Lactic acid was greatly alleviated in A2780 cells with SNHG22 abalation (Fig. 3D). Conversely, pCDNA-SNHG22 predominantly elevated ECAR (Fig. 3E), and lactic acid production (Fig. 3F) in ES-2 cells.
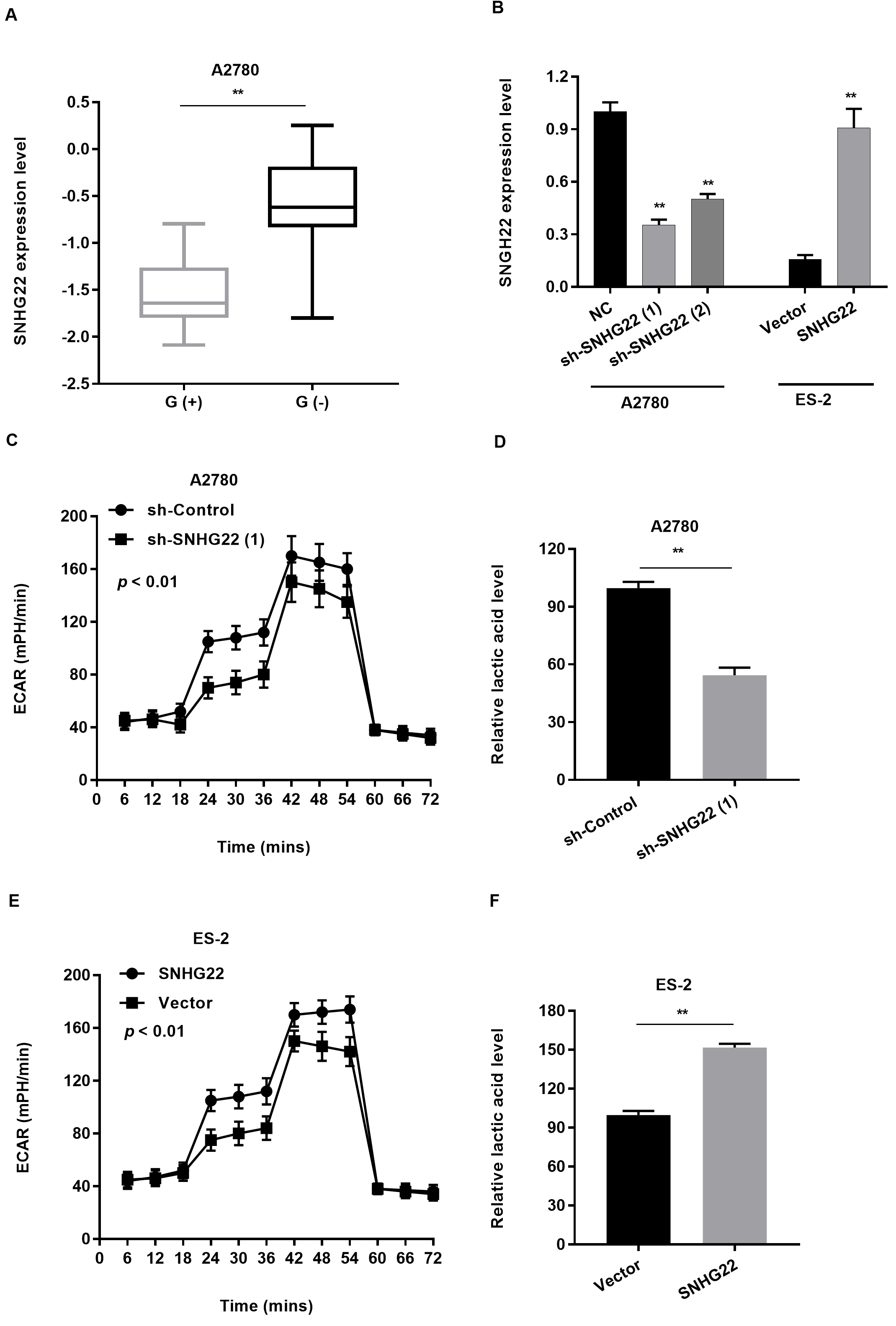 Fig. 3.
Fig. 3.SNHG22 regulates glycolysis in cancer cells.
(A) qRT-PCR analysis was used to evaluate SNHG22 expression levels in ES-2 cells
of glucose starvation.
(B) Relative expression levels of SNHG22 in transfected cells.
(C) The change of ECAR level was detected in A2780 cells transfected with
sh-control or sh-SNHG22.
(D) The relative lactic acid level was measured in A2780 cells transfected with
sh-control or sh-SNHG22.
(E) The change of ECAR level was detected in ES-2 cells transfected with vector
or pCDNA-SNHG22.
(F) The relative lactic acid level was measured in ES-2 cells transfected with
vector or pCDNA-SNHG22.
*P
We next explored the upstream regulation mechanism of SNHG22. We assumed that SP1 contributed to the upregulation of SNHG22 by binding to the promoter, according to the JASPAR and UCSC website analysis. To verify this speculation, we first suppressed the SP1 expression using siRNA (Fig. 4A) and found SNHG22 with diminished level (Fig. 4B). Second, luciferase reporter assay denoted that the SP1 depletion could weaken the SNHG22 promoter activity, suggesting that SP1 modified SNHG22 expression at the transcriptional level (Fig. 4C). Third, we explored the enrichment of the DNA motif of SP1 and the promoter region of SNHG22. In detail, we divided the promoter region into four sections on the basis of possible binding sites (Fig. 4D). And ChIP analysis showed that the enrichment of SP1 binding was obvious in the P1 promoter region (-500~1 nt) (Fig. 4E). In addition, we assembled pGL3-SNHG22 promoter full region and P1 deleted region and carried out ChIP analysis again (Fig. 4F). Intriguingly, we found that P1 section (-500~1 nt) was necessary for SP1 binding, for there is nearly no enrichment in P1 deleted group (Fig. 4G). Finally, the correlation between SP1 expression in serum and clinicopathologic characteristics of EOC patients in Supplementary Table 6.
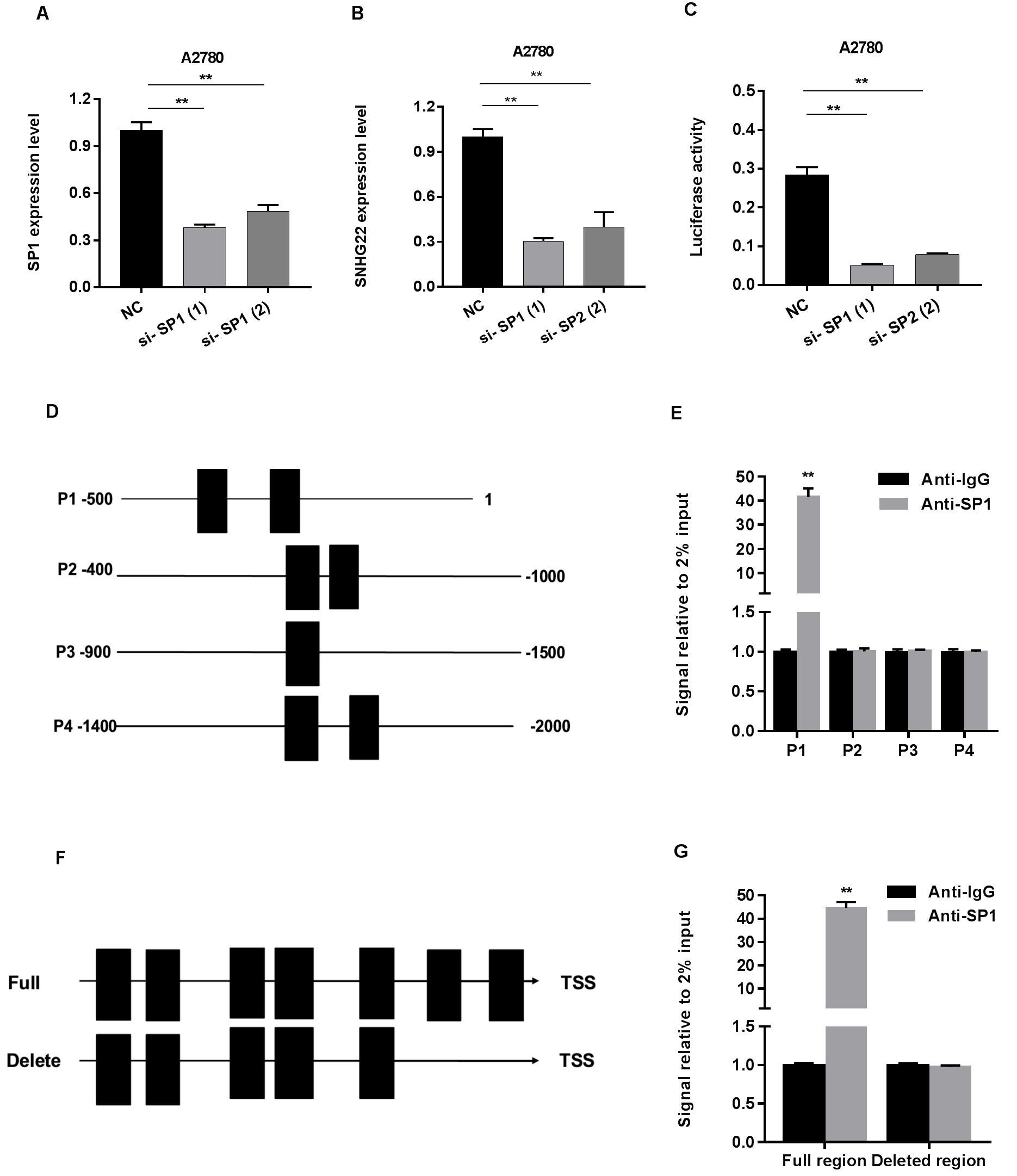 Fig. 4.
Fig. 4.SP1 regulates SNHG22 expression by serving as a transcription activator.
(A) The SP1 interference efficiency using 2 different siRNAs and negative
control detected by qRT-PCR.
(B) RT-qPCR analysis of SNHG22 expression after SP1 knockdown in A2780.
(C) Luciferase reporter assay was performed using vector-containing sequences of
the SNHG22 promoter region.
(D) SNHG22 promoter was divided into four sectional fragments.
(E) ChIP analysis was performed to detect the affinity of SP1 with SNHG22
promoter.
(F) The full LINC00958 promoter region and P1 deleted region was constructed.
(G) ChIP analysis was performed to determine the relative enrichment of SNHG22
promoter.
*P
SNHG22 was first identified in a custom annotation pipeline of micro-array data in a total of 217 Binet A CLL patients with early stage and 26 different normal B-cells [16]. It is reported that SNHG22 is an oncogene in HCC progression and is useful for biomarker development [17]. Recently, SNHG22 facilitated cell growth and motility and might serve as a new promising strategy for triple-negative breast cancer treatment [18]. Previously, SNHG22 expression had a relationship with EOC [19], but it is still unclear whether SNHG22 expression could be served as a biomarker or has a pathogenic role in glycolysis of EOC. Firstly, we verified that SNHG22 was highly expressed in a total of 60 paired tumor and non-tumor tissues and 4 EOC cell lines. Next, we discriminated the patients with EOC from healthy controls according to SNHG22 expression, with an AUC of 0.824 (95% CI: 0.747–0.900). Then our data showed that SNHG22 expression in EOC serum was significantly associated with clinicopathological features and shorter survival time. Another, our findings clarified SNHG22 expression was elevated in early and late stage in serum of patients with EOC. Besides, multivariate analysis confirmed that SNHG22 expression in serum might be an independent prognostic factor. All in all, our findings proposed that SNHG22 is an promising diagnostic and prognostic biomarker for patients with EOC.
Malignant tumors could undergo glycolysis at a higher speed than that of non-tumor tissue controls [20, 21, 22]. This phenomenon was first described in the 1920s by Otto Warburg, and is known as the Warburg effect [23]. The Warburg hypothesis demonstrates that cancer is fundamentally caused by mitochondrial metabolism disorder. Doherty JR, et al. [24] found that tumor lactate levels correlate with increased metastasis, tumor recurrence, and poor outcome. And targeting lactate metabolism is a promising approach for cancer therapeutics. Furthermore, cancer cells with high level of glycolysis and acid resistance have a energetic growth advantage, which facilitates unrestrained proliferation and invasion. In our study, our research demonstrated that SNHG22 regulates specific genes linked with metabolic pathways and SNHG22 promotes glycolysis in EOC cells. Nowadays, ketogenic diet was used to constrain glycolysis to starve cancer cells, adjusting mitochondrial metabolism [25, 26]. Herein, we will dedicate ourselves to further explore the mechanism of SNHG22 modulating glycolysis.
Transcription factor could promote or repress cancer progression via altering the expression profile of genes. SP1 is a famous transcription factor. For example, SP1 could interact with lncRNA LINC00958 promoter region and facilitate its transcription [27]. And SP1 could enrich to the core promoter region of LINC00152 in vitro and in vivo, and regulates its expression [28]. In our study, we elucidated that the upregulation of SNNHG22 was attributed to SP1 which served as a transcription activator in EOC. Hence, this research definitely provides a meaningful revelation for the critical role of transcription factor in regulating biological activities, and ectopic lncRNAs expression in malignant tumors.
Nevertheless, there are still some aspects need us to state. First, we should test the SNHG22 expression level under hypoxia, for hypoxia or the transcription factor hypoxiainducible factor 1 (HIF-1) might have effect on SNHG22 expression. Second, lncRNAs always binds protein to exert biological function, we should perform RNA pulldown and mass spectrometry to determine the SNHG22 potential binding protein.
In conclusion, our findings demonstrated that elevated SNHG22 facilitates glycolysis in EOC, which is regulated by SP1. And SNHG22 is a potential biomarker for EOC diagnosis and even useful for monitoring the recurrence of tumors by measuring the serum level after surgical resection.
XJT contributed to the conception of the study. XYL contributed significantly to analysis and manuscript preparation. LD performed the data analyses and wrote the manuscript. XLW helped perform the analysis with constructive discussions.
Patients have given their written informed consent in our study. And ethics committee of the Second People’s Hospital of Baise and liaoning cancer hospital approved the study protocol (reference number: 2015 06 25 181251).
Thank you for Helen’s work for statistic analysis.
Natural Science Foundation of Liaoning Province, Project name: Research on the role of TRIM59 in ovarian cancer cell invasion and metastasis and its mechanism. Project No.: 2019-ZD-0588. Shenyang Science and Technology Project: Research on the mechanism of TRIM59 mediating ovarian cancer cell apoptosis by activating p38MAPK pathway. Project No.: 201170.
The authors declare no conflict of interest.
